p53 domains: suppression, transformation, and transactivation
- PMID: 8508031
- PMCID: PMC6081624
p53 domains: suppression, transformation, and transactivation
Abstract
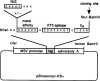
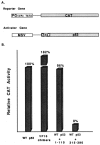
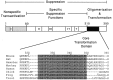
We investigated the suppression, transformation, and transactivation functions of isolated segments of wild-type murine p53. Intact p53, but no segment of p53, inhibited cellular transformation by the activated ras and adenovirus E1A proteins. We conclude that most of p53 is needed for suppression of cellular proliferation. Nevertheless, the transactivating domain of herpesvirus protein VP16 was able to substitute for the N-terminal transactivating domain of p53 in cellular suppression. Thus, unless the interchanged p53 and VP16 acidic segments share additional functions, transactivation is required for suppression by p53. Interestingly, we found that all p53 segments containing amino acids 320-360 enhanced transformation by ras and E1A. This region has been associated with the oligomerization of p53 (Milner et al., 1991; Sturzbecher et al., 1992). Furthermore, no p53 segment lacking amino acids 320-360 transformed cells. Amino acids 320-360, therefore, may account for the major transforming activity of p53. Intact p53 and chimeric VP16-p53 transactivated the CAT gene under control of a p53-specific promoter, while transforming segments of p53 interfered with transactivation by wild-type p53. Our findings argue that transactivation by p53 is required for cellular suppression and that any nontransactivating p53 that retains the capacity to oligomerize with wild-type p53 would have transformation potential.
Figures




References
-
- Baker S. J., Fearon E. R., Nigro J. M., Hamilton S. R., Preisinger A. D., Jessup J. M., vanTuinen P., Ledbetter D. H., Barker D. F., Nakamura Y., White R., and Vogelstein B. (1989), Science 244, 217–221. - PubMed
-
- Baker S. J., Markowitz S., Fearon E. R., Willson J. K. V., and Vogelstein B. (1990), Science 249, 912–915. - PubMed
-
- Bargonetti J., Friedman P. N., Kern S. E., Vogelstein B., and Prives C. (1991), Cell 65, 1083–1091. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
