Role of mutant CFTR in hypersusceptibility of cystic fibrosis patients to lung infections
- PMID: 8539601
- PMCID: PMC3677515
- DOI: 10.1126/science.271.5245.64
Role of mutant CFTR in hypersusceptibility of cystic fibrosis patients to lung infections
Abstract
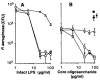
Cystic fibrosis (CF) patients are hypersusceptible to chronic Pseudomonas aeruginosa lung infections. Cultured human airway epithelial cells expressing the delta F508 allele of the cystic fibrosis transmembrane conductance regulator (CFTR) were defective in uptake of P. aeruginosa compared with cells expressing the wild-type allele. Pseudomonas aeruginosa lipopolysaccharide (LPS)-core oligosaccharide was identified as the bacterial ligand for epithelial cell ingestion; exogenous oligosaccharide inhibited bacterial ingestion in a neonatal mouse model, resulting in increased amounts of bacteria in the lungs. CFTR may contribute to a host-defense mechanism that is important for clearance of P. aeruginosa from the respiratory tract.
Figures
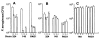
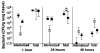
References
-
-
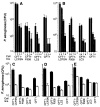
Cells were released from monolayers in tissue culture flasks by 5-min incubation with a trypsin-versene mixture (BioWhitaker, Walkersville, MD) and washed, counted, and seeded into 96-well tissue culture plates at a concentration of 105 cells per well in supplemented F-12 medium (3). Plates were incubated at either 26° or 37°C in 5% CO2. Fresh cultures of P. aeruginosa grown overnight at 37°C on a tryptic soy agar plate were suspended in supplemented F-12 medium to prepare the bacterial inoculum; then ~106 colony-forming units (CFU; range from 6 × 105 to 2.3 × 106 CFU) were added per well of 105 epithelial cells. Bacteria were allowed to be ingested by the epithelial cells for 3 or 4 hours at either 26° or 37°C, after which nonadherent bacteria were removed by washing. The remainder of the assay and controls were as described [S. M. J. Fleiszig, T. S. Zaidi, G. B. Pier, Infect. Immun. 63, 4072 (1995)]. Three to nine replicates were obtained per point and were analyzed by analysis of variance (ANOVA) and the Fisher PLSD statistic to determine pair-wise differences.
-
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

