Conserved features in papillomavirus and polyomavirus capsids
- PMID: 8656427
- PMCID: PMC4142696
- DOI: 10.1006/jmbi.1996.0317
Conserved features in papillomavirus and polyomavirus capsids
Abstract
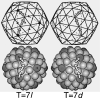
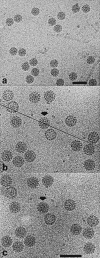
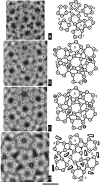
Capsids of papilloma and polyoma viruses (papovavirus family) are composed of 72 pentameric capsomeres arranged on a skewed icosahedral lattice (triangulation number of seven, T = 7). Cottontail rabbit papillomavirus (CRPV) was reported previously to be a T = 7laevo (left-handed) structure, whereas human wart virus, simian virus 40, and murine polyomavirus were shown to be T = 7dextro (right-handed). The CRPV structure determined by cryoelectron microscopy and image reconstruction was similar to previously determined structures of bovine papillomavirus type 1 (BPV-1) and human papillomavirus type 1 (HPV-1). CRPV capsids were observed in closed (compact) and open (swollen) forms. Both forms have star-shaped capsomeres, as do BPV-1 and HPV-1, but the open CRPV capsids are approximately 2 nm larger in radius. The lattice hands of all papillomaviruses examined in this study were found to be T = 7dextro. In the region of maximum contact, papillomavirus capsomeres interact in a manner similar to that found in polyomaviruses. Although papilloma and polyoma viruses have differences in capsid size (approximately 60 versus approximately 50 nm), capsomere morphology (11 to 12 nm star-shaped versus 8 nm barrel-shaped), and intercapsomere interactions (slightly different contacts between capsomeres), papovavirus capsids have a conserved, 72-pentamer, T = 7dextro structure. These features are conserved despite significant differences in amino acid sequences of the major capsid proteins. The conserved features may be a consequence of stable contacts that occur within capsomeres and flexible links that form among capsomeres.
Figures





References
-
- Adolph KW, Caspar DLD, Hollingshead CJ, Lattman EE, Philips WC, Murakami WT. Polyoma virion and capsid crystal structures. Science. 1979;203:1117–1120. - PubMed
-
- Adrian M, Dubochet J, Lepault J, McDowall AW. Cryo-electron microscopy of viruses. Nature. 1984;308:32–36. - PubMed
-
- Anderer FA, Schlumberger HD, Koch MA, Frank H, Eggers HJ. Structure of simian virus 40: II. symmetry and components of the virus particle. Virology. 1967;32:511–523. - PubMed
-
- Baker CC. Sequence analysis of papillomavirus genomes. In: Salzman NP, Howley PM, editors. The Papovaviridae, vol. 2, The Papillomaviruses. Plenum Press; New York: 1987. pp. 321–385.
-
- Baker TS. Cryo-electron microscopy and three-dimensional image reconstruction of icosahedral viruses. In: Megías-Megías L, Rodríquez-García MI, Ríos A, Arias JM, editors. Electron Microscopy 92: Proceedings of the 10th European Congress on Electron Microscopy held in Granada, Spain, 7-11 September 1992. Vol. 3. Secretariado de Publicaciones de la Universidad de Granada; Granada: 1992. pp. 275–279.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

