Structural analysis of substrate binding by the molecular chaperone DnaK
- PMID: 8658133
- PMCID: PMC5629921
- DOI: 10.1126/science.272.5268.1606
Structural analysis of substrate binding by the molecular chaperone DnaK
Abstract
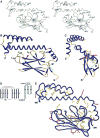
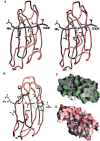
DnaK and other members of the 70-kilodalton heat-shock protein (hsp70) family promote protein folding, interaction, and translocation, both constitutively and in response to stress, by binding to unfolded polypeptide segments. These proteins have two functional units: a substrate-binding portion binds the polypeptide, and an adenosine triphosphatase portion facilitates substrate exchange. The crystal structure of a peptide complex with the substrate-binding unit of DnaK has now been determined at 2.0 angstroms resolution. The structure consists of a beta-sandwich subdomain followed by alpha-helical segments. The peptide is bound to DnaK in an extended conformation through a channel defined by loops from the beta sandwich. An alpha-helical domain stabilizes the complex, but does not contact the peptide directly. This domain is rotated in the molecules of a second crystal lattice, which suggests a model of conformation-dependent substrate binding that features a latch mechanism for maintaining long lifetime complexes.
Figures


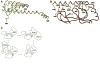
References
-
- Gething MJ, Sambrook J. Nature. 1992;355:33. - PubMed
- Hendrick JP, Hartl FU. Annu Rev Biochem. 1993;62:349. - PubMed
- Morimoto RI, Tissieres A, Georgopoulos C. In: The Biology of Heat Shock Proteins and Molecular Chaperones. Morimoto RI, Tissieres A, Georgopoulos C, editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1994.
-
- Pelham HRB. Cell. 1986;46:959. - PubMed
-
- Rothman JE. ibid. 1989;59:591. - PubMed
-
- Georgopoulos C. In: The Biology of Heat Shock Proteins and Molecular Chaperones. Morimoto RI, Tissieres A, Georgopoulos C, editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1994. pp. 209–249.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

