Identification and cloning of centaurin-alpha. A novel phosphatidylinositol 3,4,5-trisphosphate-binding protein from rat brain
- PMID: 8702546
- PMCID: PMC4298166
- DOI: 10.1074/jbc.271.31.18859
Identification and cloning of centaurin-alpha. A novel phosphatidylinositol 3,4,5-trisphosphate-binding protein from rat brain
Abstract
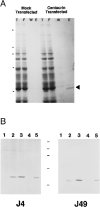
Using an affinity resin and photoaffinity label based on phospholipid analogs of inositol 1,3,4,5-tetrakisphosphate (InsP4), we have isolated, characterized, and cloned a 46-kDa protein from rat brain, which we have named centaurin-alpha. Binding specificity was determined using displacement of 1-O-[3H](3-[4-benzoyldihydrocinnamidyl]propyl)-InsP4 photoaffinity labeling. Centaurin-alpha displayed highest affinity for phosphatidylinositol 3,4,5-trisphosphate (PtdInsP3) (IC50 = 120 nM), whereas InsP4, PtdInsP2, and InsP3 bound with 5-, 12-, and >50-fold lower affinity, respectively. Screening a rat brain cDNA library with a polymerase chain reaction product, generated using partial amino acid sequence from tryptic peptides, yielded a full-length clone. The 2,450-base pair cDNA contained an open reading frame (ORF) encoding a novel protein of 419 amino acids. Northern analysis revealed a 2.5-kilobase transcript that is highly expressed in brain. The deduced sequence contains a novel putative zinc finger motif, 10 ankyrin-like repeats, and shows homology to recently identified yeast and mammalian Arf GTPase-activating proteins. Given the specificity of binding and enrichment in brain, centaurin-alpha is a candidate PtdInsP3 receptor that may link the activation of phosphoinositide 3-kinase to downstream responses in the brain.
Figures






References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

