Hydrophilic side chains in the third and seventh transmembrane helical domains of human A2A adenosine receptors are required for ligand recognition
- PMID: 8794889
- PMCID: PMC3418326
Hydrophilic side chains in the third and seventh transmembrane helical domains of human A2A adenosine receptors are required for ligand recognition
Abstract
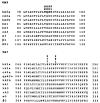
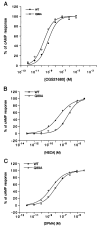
Hydrophilic residues of the G protein-coupled human A2A adenosine receptor that are potentially involved in the binding of the ribose moiety of adenosine were targeted for mutagenesis. Residues in a T88QSS91 sequence in the third transmembrane helical domain (TM3) were individually replaced with alanine and other amino acids. Two additional serine residues in TM7 that were previously shown to be involved in ligand binding were mutated to other uncharged, hydrophilic amino acids. The binding affinity of agonists at T88 mutant receptors was greatly diminished, although the receptors were well expressed and bound antagonists similar to the wild-type receptor. Thus, mutations that are specific for diminishing the affinity of ribose-containing ligands (i.e., adenosine agonists) have been identified in both TM3 and TM7. The T88A and T88S mutant receptor fully stimulated adenylyl cyclase, with the dose-response curves to CGS 21680 highly shifted to the right. A Q89A mutant gained affinity for all agonist and antagonist ligands examined in binding and functional assays. Q89 likely plays an indirect role in ligand binding. S90A, S91A, and S277C mutant receptors displayed only moderate changes in ligand affinity. A S281N mutant gained affinity for all adenosine derivatives (agonists), but antagonist affinity was generally diminished, with the exception of a novel tetrahydrobenzothiophenone derivative.
Figures
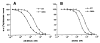


References
-
- Libert F, Parmentier M, Lefort A, Dinsart C, van Sande J, Maenhaut C, Simons MJ, Dumont JE, Vassart G. Selective amplification and cloning of four new members of the G protein-coupled receptor family. Science (Washington D. C.) 1989;244:569–572. - PubMed
-
- Jacobson M. Molecular biology of adenosine receptors. In: Belardinelli L, Pelleg A, editors. Ad.snosine and Adenine Nucleotides: From Molecular Biology to Integrative Physiology. Kluver; Norwell, MA: 1995. pp. 5–14.
-
- Barraco RA, Martens K, Parizon M, Normile HJ. Role of adenosine A2a receptors in the nucleus-accumbens. Prog. Neuropsychopharmacol. Biol. Psychiatry. 1994;18:545–553. - PubMed
-
- Olsson RA, Pearson JD. Cardiovascular purinoceptors. Pharmacol. Rev. 1990;3:761–845. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
