Role of the composite glucocorticoid response element in proliferin gene expression
- PMID: 8821626
- PMCID: PMC6138011
Role of the composite glucocorticoid response element in proliferin gene expression
Abstract
A binding site for the glucocorticoid receptor in the serum-inducible proliferin gene promoter has been reported to function as a composite glucocorticoid response element when fused to a minimal promoter. We now show that this element can also act as a glucocorticoid-independent negative regulator of transcription, both as an isolated element fused to a minimal promoter and within the context of the proliferin gene promoter. Furthermore, this element is recognized by a factor in mouse fibroblast cell extracts that is distinct from the glucocorticoid receptor and from AP-1, both of which have previously been shown to be able to bind to this site. The ability of this element to repress serum-inducible proliferin promoter activity is dependent on the position of this element with respect to the adjacent serum response region, and on the activity of a positive regulatory element located further upstream in the proliferin promoter.
Figures
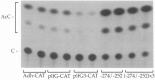


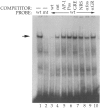


References
-
- Bradford M. M. Anal. Biochem. 72:248–254; 1976. - PubMed
-
- Diamond M. I.; Miner J. N.; Yoshinaga S. K.; Yamamoto K. R. Science 249:1266–1272; 1990. - PubMed
-
- Fienup V. K.; Jeng M. H.; Hamilton R. T.; Nilsen-Hamilton M. J. Cell. Physiol. 129:151–158; 1986. - PubMed
-
- Gametchu B.; Harrison R. W. Endocrinology 114:274–279; 1984. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
