Direct and indirect mechanisms of repression participate in suppression of T-cell-specific gene expression in T x L-cell hybrids
- PMID: 8836737
- PMCID: PMC6138021
Direct and indirect mechanisms of repression participate in suppression of T-cell-specific gene expression in T x L-cell hybrids
Abstract
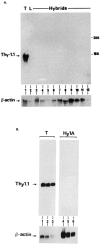
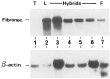
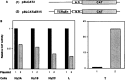
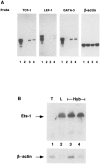
Expression of tissue-specific genes can be altered upon fusion of mammalian cells of different types. To resolve the genetic basis of this phenomenon and to identify components of the regulatory circuits that are involved, we have established a series of somatic cell hybrids between mouse T cells and L cells. These hybrids have an unusual and interesting phenotype. Unlike many hybrid cells studied, in which the expression of an entire set of tissue-specific genes was coordinately extinguished, in our T x L-cell hybrids only two out of seven T-cell-restricted genes were completely extinguished, whereas the other genes were repressed to various degrees. These hybrids extinguish the production of TCR beta and Thy-1 mRNA, repress the expression of TCR alpha, GATA-3, TCF-1, and LEF-1 genes to different extents, exhibit small changes in the level of CD3-epsilon mRNA, and continue to express the fibroblast-specific fibronectin gene, and the ets-1 gene. In this study we have evaluated for the first time the molecular mechanisms that underlie the repression of TCR alpha and TCR beta chain genes in T x L-cell hybrids. We have shown that multiple repression mechanisms, both direct and indirect, contribute to TCR alpha and TCR beta suppression. Repression of the expression of these genes correlated not only with the downregulation of GATA-3, TCF-1, and LEF-1 transcription factor expression, and with a change in the chromatin structure, but more importantly, with the activation of the silencer activity. Our study provides evidence for the existence of at least two negatively regulating elements, located at the TCR alpha enhancer-containing fragment and at the silencer region, which are active in our hybrid cells. We have shown that there was no correlation between the levels of GATA-3, TCF-1, and LEF-1 expression versus the level of TCR alpha mRNA in the independent hybrids. In contrast, both the silencer activity and the ability of the TCR alpha enhancer to downregulate thymidine kinase (TK) promoter activity were found to be in an inverse correlation with the ability of the different hybrid cells to express TCR alpha mRNA.
Figures





References
-
- Amouyel P.; Gegonne A.; Delacourte A.; Defossez A.; Stehelin D. Expression of ETS proto-oncogenes in astrocytes in human cortex. Brain Res. 447:149–153; 1988. - PubMed
-
- Ben-Shushan E.; Pikarsky E.; Klar A.; Bergman Y. Extinction of Oct-3/4 gene expression in embryonal carcinoma × fibroblast somatic cell hybrids is accompanied by changes in the methylation status, chromatin structure, and transcriptional activity of the Oct-3/4 upstream region. Mol. Cell. Biol. 13:891–901; 1993. - PMC - PubMed
-
- Bhat N. K.; Komschlies K. L.; Fujiwara S.; Fisher R. J.; Mathieson B. J.; Gregorio T. A.; Young H. A.; Kasik J. W.; Ozato K.; Papas T. S. Expression of ets genes in mouse thymocyte subsets and T cells. J. Immunol. 142:672–678; 1989. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
