Breakage in the SNRPN locus in a balanced 46,XY,t(15;19) Prader-Willi syndrome patient
- PMID: 8845846
- PMCID: PMC6057871
- DOI: 10.1093/hmg/5.4.517
Breakage in the SNRPN locus in a balanced 46,XY,t(15;19) Prader-Willi syndrome patient
Abstract
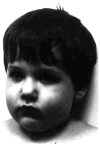
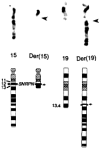
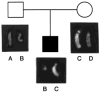
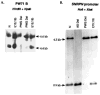
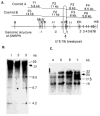
A patient with Prader-Willi syndrome (PWS) was found to carry a de novo balanced reciprocal translocation, t(15;19)(q12;q13.41), which disrupted the small nuclear ribonucleoprotein N (SNRPN) locus. The translocation chromosome 15 was found to be paternal in origin. Uniparental disomy and abnormal DNA methylation were ruled out. The translocation breakpoint was found to have occurred between exon 0 (second exon) and 1 (third exon) of the SNRPN locus outside of the SmN open reading frame (ORF), which is intact. The transcriptional activities of ZNF127, IPW, PAR-1, and PAR-5 were detected with RT-PCR from fibroblasts of the patient, suggesting that these genes may not play a significant role in the PWS phenotype in this patient. Transcription from the first two exons and last seven exons of the SNRPN gene was also detected with RT-PCR; however, the complete mRNA (10 exons) was not detected. Thus, the PWS phenotype in the patient is likely to be the result of disruption of the SNRPN locus.
Figures

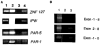

References
-
- Ledbetter DH, Riccardi VM, Airhart SD, Strobel RJ, Keenan BS, Crawford JD. Deletions of chromosome 15 as a cause of the Prader-Willi syndrome. N Engl J Med. 1981;304:325–329. - PubMed
-
- Buiting K, Saitoh S, Gross S, Dittrich B, Schwartz S, Nicholls RD, Horsthemke B. Inherited microdeletions in the Angelman and Prader-Willi syndromes define an imprinting centre on human chromosome 15. Nature Genet. 1995;9:395–400. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

