Nuclear factor YY1 activates the mammalian F0F1 ATP synthase alpha-subunit gene
- PMID: 8882641
- PMCID: PMC6138015
Nuclear factor YY1 activates the mammalian F0F1 ATP synthase alpha-subunit gene
Abstract
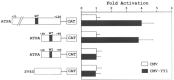
Analysis of the promoters of the bovine and human nuclear-encoded mitochondrial F0F1 ATP synthase alpha-subunit genes (ATPA) has identified several positive cis-acting regulatory regions that are important for basal promoter activity in human HeLa cells. We have previously determined that the binding of a protein factor, termed ATPF1, to an E-box sequence (CANNTG) located within one of these cis-acting regions is critical for transcriptional activation of the ATPA gene. In this article, we describe a second positive cis-acting regulatory element of the ATPA gene that is important for expression of the ATPA gene. We show that this cis-acting element also contains a binding site for a protein present in HeLa cells. On the basis of electrophoretic mobility shift patterns, oligonucleotide competition assays, and immunological cross-reactivity, we conclude that this protein factor is Yin-Yang 1 (YY1). Experiments carried out to examine the functional role of YY1 within the context of the ATPA promoter demonstrated that YY1 acts as a positive regulator of the ATPA gene. For example, when the YY1 binding site of the ATPA promoter was placed upstream of a reporter gene it was found to activate transcription in transient transfection assays. In addition, disruption of the YY1 binding site in the ATPA gene resulted in a loss of transcriptional activity. Furthermore, in cotransfection experiments overexpression of YY1 in trans was found to activate transcription of ATPA promoter-CAT constructs. Thus, at least two positive trans-acting regulatory factors, ATPF1 and YY1, are important for expression of the bovine and human F0F1 ATP synthase alpha-subunit genes.
Figures

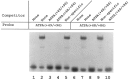
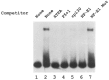
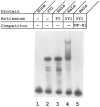

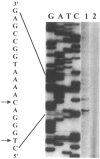

References
-
- Akiyama S.; Endo H.; Inohara N.; Ohta S.; Kagawa Y. Biochim. Biophys. Acta 1219:129–140; 1994. - PubMed
-
- Attardi G.; Schatz G. Annu. Rev. Cell Biol. 4:289–333; 1988. - PubMed
-
- Ausubel F. M.; Brent R.; Kingston R. E.; Moore D. D.; Seidman J. G.; Smith J. A.; Struhl K., eds. Current protocols in molecular biology. New York: Wiley Interscience; 1994.
-
- Basu A.; Park Y.; Atchison M. L.; Carter R. S.; Avadhani R. G. J. Biol. Chem. 268:4188–4196; 1993. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
