Pinus banksiana has at least seven expressed alcohol dehydrogenase genes in two linked groups
- PMID: 8917537
- PMCID: PMC24039
- DOI: 10.1073/pnas.93.23.13020
Pinus banksiana has at least seven expressed alcohol dehydrogenase genes in two linked groups
Abstract
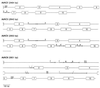
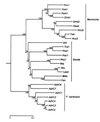
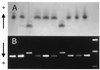
The alcohol dehydrogenase (Adh) gene family is much more complex in Pinus banksiana than in angio-sperms, with at least seven expressed genes organized as two tightly linked clusters. Intron number and position are highly conserved between P. banksiana and angiosperms. Unlike angiosperm Adh genes, numerous duplications, as large as 217 bp, were observed within the noncoding regions of P. banksiana Adh genes and may be a common feature of conifer genes. A high frequency of duplication over a wide range of scales may contribute to the large genome size of conifers.
Figures
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

