Temporal mapping of gene expression levels during the differentiation of individual primary hematopoietic cells
- PMID: 8917561
- PMCID: PMC24063
- DOI: 10.1073/pnas.93.23.13158
Temporal mapping of gene expression levels during the differentiation of individual primary hematopoietic cells
Abstract
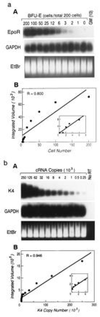
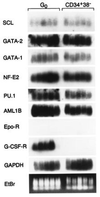
A hierarchical order of gene expression has been proposed to control developmental events in hematopoiesis, but direct demonstration of the temporal relationships between regulatory gene expression and differentiation has been difficult to achieve. We modified a single-cell PCR method to detect 2-fold changes in mRNA copies per cell (dynamic range, 250-250,000 copies/cell) and used it to sequentially quantitate gene expression levels as single primitive (CD34+,CD38-) progenitor cells underwent differentiation to become erythrocytes, granulocytes, or monocyte/macrophages. Markers of differentiation such as CD34 or cytokine receptor mRNAs and transcription factors associated with their regulation were assessed. All transcription factors tested were expressed in multipotent progenitors. During lineage-specific differentiation, however, distinct patterns of expression emerged. SCL, GATA-2, and GATA-1 expression sequentially extinguished during erythroid differentiation. PU.1, AML1B, and C/EBP alpha expression profiles and their relationship to cytokine receptor expression in maturing granulocytes could be distinguished from similar profiles in monocytic cells. These data characterize the dynamics of gene expression accompanying blood cell development and define a signature gene expression pattern for specific stages of hematopoietic differentiation.
Figures




References
-
- Shivdasani R A, Orkin S H. Blood. 1996;87:4025–4039. - PubMed
-
- Kulessa H, Frampton J, Graf T. Genes Dev. 1995;9275:1250–1262. - PubMed
-
- Zhang D E, Hohaus S, Voso M T, Chen H M, Smith L T, Hetherington C J, Tenen D G. Curr Top Microbiol Immunol. 1996;211:137–147. - PubMed
-
- Huang S, Terstappen L W M M. Blood. 1994;83:1515–1526. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

