Cloning and characterization of postsynaptic density 93, a nitric oxide synthase interacting protein
- PMID: 8922396
- PMCID: PMC6579097
- DOI: 10.1523/JNEUROSCI.16-23-07407.1996
Cloning and characterization of postsynaptic density 93, a nitric oxide synthase interacting protein
Abstract
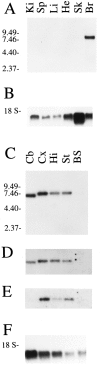
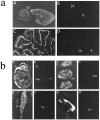
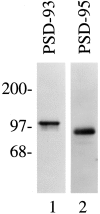
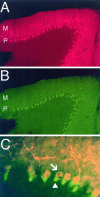
Nitric oxide (NO) formation in brain is regulated by the calcium/calmodulin dependence of neuronal NO synthase (nNOS). Calcium influx through NMDA-type glutamate receptors is efficiently coupled to nNOS activity, whereas many other intracellular calcium pathways are poorly coupled. To elucidate possible mechanisms responsible for this coupling, we performed yeast two-hybrid screening to identify proteins that interact with nNOS. Two nNOS interacting proteins were identified: the postsynaptic density proteins PSD-93 and PSD-95. Here, we report the cloning and characterization of PSD-93. PSD-93 is expressed in discrete neuronal populations as well as in specific non-neuronal cells, and it exhibits complex molecular diversity attributable to tissue-specific alternative splicing. PSD-93, like PSD-95, binds to nNOS and to the NMDA receptor 2B. PSD-93, however, is unique among PSD-95/SAP-90 family members in its expression in Purkinje neuron cell bodies and dendrites. We also demonstrate that the PDZ domain at the N terminus of nNOS is required, but it is not sufficient for interaction with PSD-93/95. Given that PSD-93 and PSD-95 each contain multiple potential binding sites for nNOS and the NMDA receptor, complexes involving oligomers of PSD-93/95 may help account for the functional as well as the physical coupling of nNOS to NMDA receptors.
Figures



References
-
- Aoki C, Fenstemaker S, Lubin M, Go CG. Nitric oxide synthase in the visual cortex of monocular monkeys as revealed by light and electron microscopic immunocytochemistry. Brain Res. 1993;620:97–113. - PubMed
-
- Bredt DS, Snyder SH. Nitric oxide, a novel neuronal messenger. Neuron. 1992;8:3–11. - PubMed
-
- Brenman JE, Chao DS, Xia H, Aldape K, Bredt DS. Nitric oxide synthase complexed with dystrophin and absent from skeletal muscle sarcolemma in Duchenne muscular dystrophy. Cell. 1995;82:743–752. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
