A helper-dependent adenovirus vector system: removal of helper virus by Cre-mediated excision of the viral packaging signal
- PMID: 8942974
- PMCID: PMC19345
- DOI: 10.1073/pnas.93.24.13565
A helper-dependent adenovirus vector system: removal of helper virus by Cre-mediated excision of the viral packaging signal
Abstract
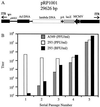
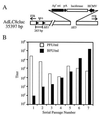
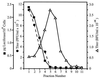
Adenoviruses are attractive vectors for the delivery of foreign genes into mammalian cells for gene therapy. However, current vectors retain many viral genes that, when expressed at low levels, contribute to the induction of a host immune response against transduced cells. We have developed a helper-dependent packaging system for production of vectors that have large regions of the genome deleted. Helper viruses were constructed with packaging signals flanked by loxP sites so that in 293 cells that stably express the Cre recombinase (293Cre), the packaging signal was efficiently excised, rendering the helper virus genome unpackageable. However, the helper virus DNA was replicated at normal levels and could thus express all of the functions necessary in trans for replication and packaging of a vector genome containing the appropriate cis-acting elements. Serial passage of the vector in helper virus-infected 293Cre cells resulted in an approximately 10-fold increase in vector titer per passage. The vector could be partially separated from residual helper virus by cesium chloride buoyant density centrifugation. Large scale preparations of vector yielded semi-purified stocks of approximately 10(10) transducing virions/ml, with < 0.01% contamination by the E1-deleted helper virus. This system should have great utility for the generation of adenovirus-based vectors with increased cloning capacity, increased safety and reduced immunogenicity.
Figures


References
-
- Hitt M, Bett A J, Addison C L, Prevec L, Graham F L. In: Methods in Molecular Genetics. Adolph K W, editor. Vol. 7. San Diego: Academic; 1995. pp. 13–30.
-
- Graham F L, Smiley J, Russell W C, Nairn R. J Gen Virol. 1977;36:59–72. - PubMed
-
- Brody S L, Crystal R G. Ann NY Acad Sci. 1994;716:90–101. - PubMed
-
- Bramson J L, Graham F L, Gauldie J. Curr Opin Biotechnol. 1995;6:590–595. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

