Evidence for a transcriptional activation function of BRCA1 C-terminal region
- PMID: 8942979
- PMCID: PMC19361
- DOI: 10.1073/pnas.93.24.13595
Evidence for a transcriptional activation function of BRCA1 C-terminal region
Abstract
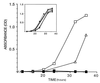
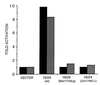
Mutations in BRCA1 account for 45% of families with high incidence of breast cancer and for 80-90% of families with both breast and ovarian cancer. BRCA1 protein includes an amino-terminal zinc finger motif as well as an excess of negatively charged amino acids near the C terminus. In addition, BRCA1 contains two nuclear localization signals and localizes to the nucleus of normal cells. While these features suggest a role in transcriptional regulation, no function has been assigned to BRCA1. Here, we show that the C-terminal region, comprising exons 16-24 (aa 1560-1863) of BRCA1 fused to GAL4 DNA binding domain can activate transcription both in yeast and mammalian cells. Furthermore, we define the region comprising exons 21-24 (aa 1760-1863) as the minimal transactivation domain. Any one of four germ-line mutations in the C-terminal region found in patients with breast or ovarian cancer (Ala-1708-->Glu, Gln-1756 C+, Met-1775-->Arg, Tyr-1853 ->Stop), had markedly impaired transcription activity. Together these data underscore the notion that one of the functions of BRCA1 may be the regulation of transcription.
Figures


References
-
- Hall J F, Lee M K, Newman B, Morrow J E, Anderson L A, Huey B, King M-C. Science. 1990;250:1684–1689. - PubMed
-
- Miki Y, Swensen J, Shattuck-Eidens D, Futreal P A, Harshman K, et al. Science. 1994;266:66–71. - PubMed
-
- Saurin A J, Borden K L B, Boddy M N, Freemont P S. Trends Biochem Sci. 1996;21:208–214. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

