hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6
- PMID: 8942985
- PMCID: PMC19374
- DOI: 10.1073/pnas.93.24.13629
hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6
Abstract
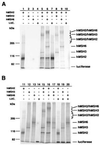
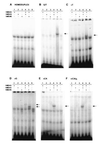
The genetic and biochemical properties of three human MutS homologues, hMSH2, hMSH3, and hMSH6, have been examined. The full-length hMSH6 cDNA and genomic locus were isolated and characterized, and it was demonstrated that the hMSH6 gene consisted of 10 exons and mapped to chromosome 2p15-16. The hMSH3 cDNA was in some cases found to contain a 27-bp deletion resulting in a loss of nine amino acids, depending on the individual from which the cDNA was isolated. hMSH2, hMSH3, and hMSH6 all showed similar tissue-specific expression patterns. hMSH2 protein formed a complex with both hMSH3 and hMSH6 proteins, similar to protein complexes demonstrated by studies of the Saccharomyces cerevisiae MSH2, MSH3, and MSH6. hMSH2 was also found to form a homomultimer complex, but neither hMSH3 nor hMSH6 appear to interact with themselves or each other. Analysis of the mismatched nucleotide-binding specificity of the hMSH2-hMSH3 and hMSH2-hMSH6 protein complexes showed that they have overlapping but not identical binding specificity. These results help to explain the distribution of mutations in different mismatch-repair genes seen in hereditary nonpolyposis colon cancer.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

