A critical arginine residue mediates cooperativity in the contact interface between transcription factors NFAT and AP-1
- PMID: 8942992
- PMCID: PMC19386
- DOI: 10.1073/pnas.93.24.13671
A critical arginine residue mediates cooperativity in the contact interface between transcription factors NFAT and AP-1
Abstract
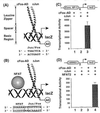
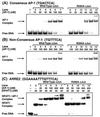
The heterologous transcription factors NFAT and AP-1 coordinately regulate cytokine gene expression through cooperative binding to precisely juxtaposed DNA recognition elements. The molecular origins of cooperativity in the binding of NFAT and AP-1 to DNA are poorly understood. Herein we have used yeast one-hybrid screening and alanine-scanning mutagenesis to identify residues in AP-1 that affect cooperative interactions with NFAT on DNA. Mutation of a single conserved Arg residue to Ala in the cJun spacer region (R285A) led to a virtually complete abolition of cooperative interactions with NFAT. The DNA-binding activity of AP-1 alone was unaffected by the cJun R285A mutation, thus indicating that this residue influences cooperative binding only. Ala-scanning mutations elsewhere in AP-1, including the cFos subunit, revealed no other strongly interacting single positions. We thus conclude that NFAT contacts AP-1 in the spacer region of the cJun subunit, making an especially important contact to R285, and that these interactions drive formation of the cooperative NFAT/AP-1/DNA complex. These results provide a general strategy for selectively ablating cooperativity between transcription factors without affecting their ability to act alone and yield insights into the structural basis for coordinate regulation of gene expression.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

