Metal binding properties and secondary structure of the zinc-binding domain of Nup475
- PMID: 8943007
- PMCID: PMC19415
- DOI: 10.1073/pnas.93.24.13754
Metal binding properties and secondary structure of the zinc-binding domain of Nup475
Abstract
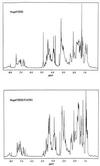
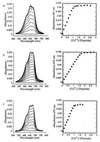
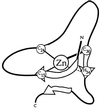
Nup475 is a nuclear zinc-binding protein of unknown function that is induced in mammalian cells by growth factor mitogens. Nup475 contains two tandemly repeated sequences YKTELCX8CX5CX3H (Cys3His repeats) that are thought to be zinc-bindin domains. Similar sequences have been found in a number of proteins from various species of eukaryotes. To determine the metal binding properties and secondary structure of the putative zinc-binding domains of Nup475, we have used synthetic or recombinant peptides that contain one or two domain sequences. The peptide with a single domain bound 1.0 +/- 0.1 equivalents of Co2+, and the peptide with two domains bound 1.7 +/- 0.4 equivalents of Co2+. Both peptides bound Co2+ and Zn2+ with affinities similar to those of classical zinc finger peptides. In each case, the Co2+ complex exhibited strong d-d transitions characteristic of tetrahedral coordination. For structural studies by nuclear magnetic resonance spectroscopy, we used a more soluble two-domain peptide that had a single amino acid substitution in a nonconserved amino acid residue in the second Cys3His repeat. The mutant peptide unexpectedly showed loss of one of its metal binding sites and displayed ordered structure for only the first Cys3His sequence. On the basis of the nuclear magnetic resonance data, we propose a structure for the Nup475 metal-binding domain in which the zinc ion is coordinated by the conserved cysteines and histidine, and the conserved YKTEL motif forms a parallel sheet-like structure with the C terminus of this domain. This structure is unlike that of any previously described class of metal binding domain.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

