Cell cycle-regulated generation of single-stranded G-rich DNA in the absence of telomerase
- PMID: 8943033
- PMCID: PMC19463
- DOI: 10.1073/pnas.93.24.13902
Cell cycle-regulated generation of single-stranded G-rich DNA in the absence of telomerase
Abstract
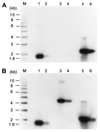
Current models of telomere replication predict that due to the properties of the polymerases implicated in semiconservative replication of linear DNA, the two daughter molecules have one end that is blunt and one end with a short 3' overhang. Telomerase is thought to extend the short 3' overhang to produce long single-stranded overhangs. Recently, such overhangs, or TG1-3 tails, were shown to occur on both telomeres of replicated linear plasmids in yeast. Moreover, indirect evidence suggested that the TG1-3 tails also occurred in a yeast strain lacking telomerase. We report herein a novel in-gel hybridization technique to probe telomeres for single-stranded DNA. Using this method, it is shown directly that in yeast strains lacking the TLC1 gene encoding the yeast telomerase RNA, TG1-3 single-stranded DNA was generated on chromosomal and plasmid telomeres. The single-stranded DNA only appeared in S phase and was sensitive to digestion with a single-strand-specific exonuclease. These data demonstrate that during replication of telomeres, TG1-3 tails can be generated in a way that is independent of telomerase-mediated strand elongation. In wild-type strains, these TG1-3 tails could subsequently serve as substrates for telomerase and telomere binding proteins on all telomeres.
Figures


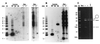
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

