Transcription-induced mutations: increase in C to T mutations in the nontranscribed strand during transcription in Escherichia coli
- PMID: 8943036
- PMCID: PMC19468
- DOI: 10.1073/pnas.93.24.13919
Transcription-induced mutations: increase in C to T mutations in the nontranscribed strand during transcription in Escherichia coli
Abstract
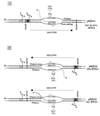
Cytosines in single-stranded DNA deaminate to uracils at 140 times the rate for cytosines in double-stranded DNA. If resulting uracils are not replaced with cytosine, C to T mutations occur. These facts suggest that cellular processes such as transcription that create single-stranded DNA should promote C to T mutations. We tested this hypothesis with the Escherichia coli tac promoter and found that induction of transcription causes approximately 4-fold increase in the frequency of C to U or 5-methylcytosine to T deaminations in the nontranscribed strand. Excess mutations caused by C to U deaminations were reduced, but not eliminated, by uracil-DNA glycosylase. Similarly, mutations caused by 5-methylcytosine to T deaminations were only partially reduced by the very short-patch repair process in E.coli. These effects are unlikely to be caused by differential repair of the two strands, and our results suggest that all actively transcribed genes in E. coli should acquire more C to T mutations in the nontranscribed strand.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

