Crystal structure of the breakpoint cluster region-homology domain from phosphoinositide 3-kinase p85 alpha subunit
- PMID: 8962058
- PMCID: PMC26139
- DOI: 10.1073/pnas.93.25.14373
Crystal structure of the breakpoint cluster region-homology domain from phosphoinositide 3-kinase p85 alpha subunit
Abstract
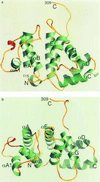
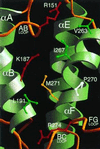
Proteins such as the product of the break-point cluster region, chimaerin, and the Src homology 3-binding protein 3BP1, are GTPase activating proteins (GAPs) for members of the Rho subfamily of small GTP-binding proteins (G proteins or GTPases). A 200-residue region, named the breakpoint cluster region-homology (BH) domain, is responsible for the GAP activity. We describe here the crystal structure of the BH domain from the p85 subunit of phosphatidylinositol 3-kinase at 2.0 A resolution. The domain is composed of seven helices, having a previously unobserved arrangement. A core of four helices contains most residues that are conserved in the BH family. Their packing suggests the location of a G-protein binding site. This structure of a GAP-like domain for small GTP-binding proteins provides a framework for analyzing the function of this class of molecules.
Figures



References
-
- Bourne H R, Sanders D A, McCormick F. Nature (London) 1991;349:117–127. - PubMed
-
- Ridley A J. J Cell Sci Suppl. 1994;18:127–131. - PubMed
-
- Hall A. Cell. 1992;69:389–391. - PubMed
-
- Vojtek A B, Cooper J A. Cell. 1995;82:527–529. - PubMed
-
- Boguski M S, McCormick F. Nature (London) 1993;366:643–654. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

