Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels
- PMID: 8962070
- PMCID: PMC26151
- DOI: 10.1073/pnas.93.25.14440
Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels
Abstract
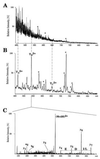
The function of many of the uncharacterized open reading frames discovered by genomic sequencing can be determined at the level of expressed gene products, the proteome. However, identifying the cognate gene from minute amounts of protein has been one of the major problems in molecular biology. Using yeast as an example, we demonstrate here that mass spectrometric protein identification is a general solution to this problem given a completely sequenced genome. As a first screen, our strategy uses automated laser desorption ionization mass spectrometry of the peptide mixtures produced by in-gel tryptic digestion of a protein. Up to 90% of proteins are identified by searching sequence data bases by lists of peptide masses obtained with high accuracy. The remaining proteins are identified by partially sequencing several peptides of the unseparated mixture by nanoelectrospray tandem mass spectrometry followed by data base searching with multiple peptide sequence tags. In blind trials, the method led to unambiguous identification in all cases. In the largest individual protein identification project to date, a total of 150 gel spots-many of them at subpicomole amounts-were successfully analyzed, greatly enlarging a yeast two-dimensional gel data base. More than 32 proteins were novel and matched to previously uncharacterized open reading frames in the yeast genome. This study establishes that mass spectrometry provides the required throughput, the certainty of identification, and the general applicability to serve as the method of choice to connect genome and proteome.
Figures

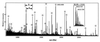

References
-
- Goffeau A, Barrell B G, Bussey H, Davis R W, Dujon B, Feldmann H, Galibert F, Hoheisal J D, Jacq L, Johnston M, Louis E J, Mewes H W, Murakami Y, Philippsen P, Tettelin H, Oliver S B. Science. 1996;274:546–567. - PubMed
-
- Wilkins M R, Pasquali C, Appel R D, Ou K, Golaz O, Sanchez J C, Yan J X, Gooley A A, Hughes G, Humphery-Smith I, Williams K L, Hochstrasser D F. Bio/Technology. 1996;14:61–65. - PubMed
-
- Kahn P. Science. 1995;270:369–370. - PubMed
-
- Mann M, Højrup P, Roepstorff P. Biol Mass Spectrom. 1993;22:338–345. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

