Dominant negative inhibition by fragments of a monomeric enzyme
- PMID: 8962072
- PMCID: PMC26153
- DOI: 10.1073/pnas.93.25.14452
Dominant negative inhibition by fragments of a monomeric enzyme
Abstract
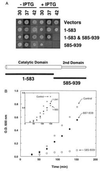
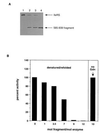
Dominant negative inhibition is most commonly seen when a mutant subunit of a multisubunit protein is coexpressed with the wild-type protein so that assembly of a functional oligomer is impaired. By analogy, it should be possible to interfere with the functional assembly of a monomeric enzyme by interfering with the folding pathway. Experiments in vitro by others suggested that fragments of a monomeric enzyme might be exploited for this purpose. We report here dominant negative inhibition of bacterial cell growth by expression of fragments of a tRNA synthetase. Inhibition is fragment-specific, as not all fragments cause inhibition. An inhibitory fragment characterized in more detail forms a specific complex with the intact enzyme in vivo, leading to enzyme inactivation. This fragment also associated stoichiometrically with the full-length enzyme in vitro after denaturation and refolding, and the resulting complex was catalytically inactive. Inhibition therefore appears to arise from an interruption in the folding pathway of the wild-type enzyme, thus suggesting a new strategy to design dominant negative inhibitors of monomeric enzymes.
Figures


References
-
- Herskowitz I. Nature (London) 1987;329:219–222. - PubMed
-
- Amaya E, Musci T J, Kirschner M W. Cell. 1991;66:257–270. - PubMed
-
- Verrall S, Hall Z W. Cell. 1992;68:23–31. - PubMed
-
- Kintner C. Cell. 1992;69:225–236. - PubMed
-
- Friedman A D, Triezenberg S J, McKnight S L. Nature (London) 1988;335:452–454. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

