Molecular ordering of the Fas-apoptotic pathway: the Fas/APO-1 protease Mch5 is a CrmA-inhibitable protease that activates multiple Ced-3/ICE-like cysteine proteases
- PMID: 8962078
- PMCID: PMC26159
- DOI: 10.1073/pnas.93.25.14486
Molecular ordering of the Fas-apoptotic pathway: the Fas/APO-1 protease Mch5 is a CrmA-inhibitable protease that activates multiple Ced-3/ICE-like cysteine proteases
Abstract
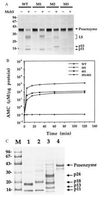
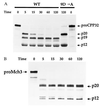
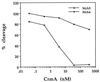
The Fas/APO-1-receptor associated cysteine protease Mch5 (MACH/FLICE) is believed to be the enzyme responsible for activating a protease cascade after Fas-receptor ligation, leading to cell death. The Fas-apoptotic pathway is potently inhibited by the cowpox serpin CrmA, suggesting that Mch5 could be the target of this serpin. Bacterial expression of proMch5 generated a mature enzyme composed of two subunits, which are derived from the pre-cursor proenzyme by processing at Asp-227, Asp-233, Asp-391, and Asp-401. We demonstrate that recombinant Mch5 is able to process/activate all known ICE/Ced-3-like cysteine proteases and is potently inhibited by CrmA. This contrasts with the observation that Mch4, the second FADD-related cysteine protease that is also able to process/activate all known ICE/Ced-3-like cysteine proteases, is poorly inhibited by CrmA. These data suggest that Mch5 is the most upstream protease that receives the activation signal from the Fas-receptor to initiate the apoptotic protease cascade that leads to activation of ICE-like proteases (TX, ICE, and ICE-relIII), Ced-3-like proteases (CPP32, Mch2, Mch3, Mch4, and Mch6), and the ICH-1 protease. On the other hand, Mch4 could be a second upstream protease that is responsible for activation of the same protease cascade in CrmA-insensitive apoptotic pathways.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

