Development of an in vitro mRNA decay system for Escherichia coli: poly(A) polymerase I is necessary to trigger degradation
- PMID: 8975250
- PMCID: PMC24022
- DOI: 10.1073/pnas.93.23.12926
Development of an in vitro mRNA decay system for Escherichia coli: poly(A) polymerase I is necessary to trigger degradation
Abstract
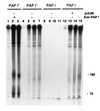
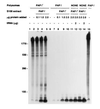
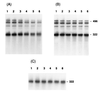
Using a novel Escherichia coli in vitro decay system in which polysomes are the source of both enzymes and mRNA, we demonstrate a requirement for poly(A) polymerase I (PAP I) in mRNA turnover. The in vitro decay of two different mRNAs (trxA and lpp) is triggered by the addition of ATP only when polysomes are prepared from s strain carrying the wild-type gene for PAP I (pcnB+). The relative decay rates of these two messages are similar in vitro and in vivo. Poly(A) tails are formed on both mRNAs, but no poly(A) are detected on the 3' end of mature 23S rRNA. The size distribution of poly(A) tails generated in vitro, averaging 50 nt in length, is comparable to that previously reported in vivo. PAP I activity is associated exclusively with the polysomes. Exogenously added PAP I does not restore mRNA decay to PAP I-polysomes, suggesting that, in vivo, PAP I may be part of a multiprotein complex. The potential of this in vitro system for analyzing mRNA decay in E. coli is discussed.
Figures




Similar articles
-
Polyadenylylation helps regulate mRNA decay in Escherichia coli.Proc Natl Acad Sci U S A. 1995 Mar 14;92(6):1807-11. doi: 10.1073/pnas.92.6.1807. Proc Natl Acad Sci U S A. 1995. PMID: 7534403 Free PMC article.
-
Analysis of the function of Escherichia coli poly(A) polymerase I in RNA metabolism.Mol Microbiol. 1999 Dec;34(5):1094-108. doi: 10.1046/j.1365-2958.1999.01673.x. Mol Microbiol. 1999. PMID: 10594833
-
Identification of a second poly(A) polymerase in Escherichia coli.Biochem Biophys Res Commun. 1994 Jan 28;198(2):459-65. doi: 10.1006/bbrc.1994.1067. Biochem Biophys Res Commun. 1994. PMID: 7905263
-
Polyadenylation of mRNA in bacteria.Microbiology (Reading). 1996 Nov;142 ( Pt 11):3125-33. doi: 10.1099/13500872-142-11-3125. Microbiology (Reading). 1996. PMID: 8969510 Review. No abstract available.
-
Surprises at the 3' end of prokaryotic RNA.Cell. 1995 Mar 24;80(6):829-32. doi: 10.1016/0092-8674(95)90284-8. Cell. 1995. PMID: 7535193 Review. No abstract available.
Cited by
-
Unpaired terminal nucleotides and 5' monophosphorylation govern 3' polyadenylation by Escherichia coli poly(A) polymerase I.Proc Natl Acad Sci U S A. 2000 Jun 6;97(12):6415-20. doi: 10.1073/pnas.120173797. Proc Natl Acad Sci U S A. 2000. PMID: 10823925 Free PMC article.
-
The RNA binding protein Hfq interacts specifically with tRNAs.RNA. 2008 Mar;14(3):514-23. doi: 10.1261/rna.531408. Epub 2008 Jan 29. RNA. 2008. PMID: 18230766 Free PMC article.
-
The Polyadenylation of RNA in Plants.Plant Physiol. 1997 Oct;115(2):321-325. doi: 10.1104/pp.115.2.321. Plant Physiol. 1997. PMID: 12223809 Free PMC article. No abstract available.
-
Characterization of a cDNA encoding a novel plant poly(A) polymerase.Plant Mol Biol. 1998 Jul;37(4):729-34. doi: 10.1023/a:1006000213403. Plant Mol Biol. 1998. PMID: 9687075
-
A comparative analysis of two conserved motifs in bacterial poly(A) polymerase and CCA-adding enzyme.Nucleic Acids Res. 2008 Sep;36(16):5212-20. doi: 10.1093/nar/gkn494. Epub 2008 Aug 5. Nucleic Acids Res. 2008. PMID: 18682528 Free PMC article.
References
-
- Grunberg-Manago M. Prog Nucleic Acids Res. 1963;1:93–133.
-
- Nikolaev N, Folsom V, Schlessinger D. Biochem Biophys Res Commun. 1976;70:920–924. - PubMed
-
- Ghora B K, Apirion D. Cell. 1978;15:1055–1066. - PubMed
-
- Kuwano M, Ono M, Endo H, Hori K, Nakamura K, Hirota Y, Ohnishi Y. Mol Gen Genet. 1977;154:279–285. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

