Identification of fission yeast nuclear markers using random polypeptide fusions with green fluorescent protein
- PMID: 8986778
- PMCID: PMC26371
- DOI: 10.1073/pnas.93.26.15146
Identification of fission yeast nuclear markers using random polypeptide fusions with green fluorescent protein
Abstract
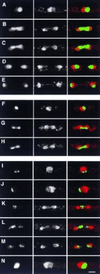
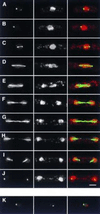
We describe a method for identifying genes encoding proteins with stereospecific intracellular localizations in the fission yeast Schizosaccharomyces pombe. Yeast are transformed with a gene library in which S. pombe genomic sequences are fused to the gene encoding the Aequorea victoria green fluorescent protein (GFP), and intracellular localizations are subsequently identified by rapid fluorescence screening in vivo. In a model application of these methods to the fission yeast nucleus, we have identified several novel genes whose products are found in specific nuclear regions, including chromatin, the nucleolus, and the mitotic spindle, and sequence similarities between some of these genes and previously identified genes encoding nuclear proteins have validated the approach. These methods will be useful in identifying additional components of the S. pombe nucleus, and further extensions of this approach should also be applicable to a more comprehensive identification of the elements of intracellular architecture in fission yeast.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

