Molecular recognition of pathogen attack occurs inside of plant cells in plant disease resistance specified by the Arabidopsis genes RPS2 and RPM1
- PMID: 8986840
- PMCID: PMC26433
- DOI: 10.1073/pnas.93.26.15497
Molecular recognition of pathogen attack occurs inside of plant cells in plant disease resistance specified by the Arabidopsis genes RPS2 and RPM1
Abstract
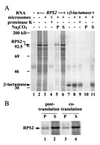
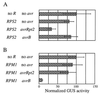
The Arabidopsis thaliana disease resistance genes RPS2 and RPM1 belong to a class of plant disease resistance genes that encode proteins that contain an N-terminal tripartite nucleotide binding site (NBS) and a C-terminal tandem array of leucine-rich repeats. RPS2 and RPM1 confer resistance to strains of the bacterial phytopathogen Pseudomonas syringae carrying the avirulence genes avrRpt2 and avrB, respectively. In these gene-for-gene relationships, it has been proposed that pathogen avirulence genes generate specific ligands that are recognized by cognate receptors encoded by the corresponding plant resistance genes. To test this hypothesis, it is crucial to know the site of the potential molecular recognition. Mutational analysis of RPS2 protein and in vitro translation/translocation studies indicated that RPS2 protein is localized in the plant cytoplasm. To determine whether avirulence gene products themselves are the ligands for resistance proteins, we expressed the avrRpt2 and avrB genes directly in plant cell using a novel quantitative transient expression assay, and found that expression of avrRpt2 and avrB elicited a resistance response in plants carrying the corresponding resistance genes. This observation indicates that no bacterial factors other than the avirulence gene products are required for the specific resistance response as long as the avirulence gene products are correctly localized. We propose that molecular recognition of P. syringae in RPS2- and RPM1-specified resistance occurs inside of plant cells.
Figures



References
-
- Keen N T. Plant Mol Biol. 1992;19:109–122. - PubMed
-
- Lamb C J, Lawton M A, Dron M, Dixon R A. Cell. 1989;56:215–224. - PubMed
-
- Flor H H. Annu Rev Phytopathol. 1971;9:275–296.
-
- Gabriel D W, Rolfe B G. Annu Rev Phytopathol. 1990;28:365–391.
-
- Staskawicz B J, Ausubel F M, Baker B J, Ellis J G, Jones J D G. Science. 1995;268:661–667. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

