The type 1 inositol 1,4,5-trisphosphate receptor gene is altered in the opisthotonos mouse
- PMID: 8987786
- PMCID: PMC6573232
- DOI: 10.1523/JNEUROSCI.17-02-00635.1997
The type 1 inositol 1,4,5-trisphosphate receptor gene is altered in the opisthotonos mouse
Abstract
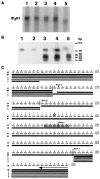
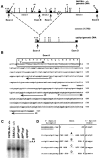
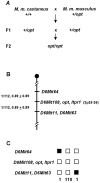
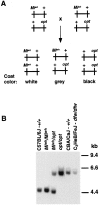
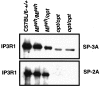
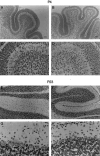
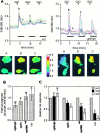
The opisthotonos (opt) mutation arose spontaneously in a C57BL/Ks-db2J colony and is the only known, naturally occurring allele of opt. This mutant mouse was first identified based on its ataxic and convulsive phenotype. Genetic and molecular data presented here demonstrate that the type 1 inositol 1,4,5-trisphosphate receptor (IP3R1) protein, which serves as an IP3-gated channel to release calcium from intracellular stores, is altered in the opt mutant. A genomic deletion in the IP3R1 gene removes two exons from the IP3R1 mRNA but does not interrupt the translational reading frame. The altered protein is predicted to have lost several modulatory sites and is present at markedly reduced levels in opt homozygotes. Nonetheless, a strong calcium release from intracellular stores can be elicited in cerebellar Purkinje neurons treated with the metabotropic glutamate receptor (mGluR) agonist quisqualate (QA). QA activates Group 1 mGluRs linked to GTP-binding proteins that stimulate phospholipase C and subsequent production of the intracellular messenger IP3, leading to calcium mobilization via the IP3R1 protein. The calcium response in opt homozygotes shows less attenuation to repeated QA application than in control littermates. These data suggest that the convulsions and ataxia observed in opt mice may be caused by the physiological dysregulation of a functional IP3R1 protein.
Figures
References
-
- Altman J. Postnatal development of the cerebellar cortex in the rat. J Comp Neurol. 1972;145:399–464. - PubMed
-
- Berridge MJ. Inositol trisphosphate and calcium signalling. Nature. 1993;361:315–325. - PubMed
-
- Bettler B, Boulter J, Hermans-Borgmeyer I, O’Shea-Greenfield A, Deneris ES, Moll C, Borgmeyer U, Hollmann M, Heinemann S. Cloning of a novel glutamate receptor subunit, GluR5: expression in the nervous system during development. Neuron. 1990;5:583–595. - PubMed
-
- Bezprozvanny I, Ehrlich BE. The inositol 1,4,5-trisphosphate (InsP3) receptor. J Membr Biol. 1995;145:205–216. - PubMed
-
- Cameron AM, Steiner JP, Roskams AJ, Ali SM, Ronnett GV, Snyder SH. Calcineurin associated with the inositol 1,4,5-trisphosphate receptor-FKBP12 complex modulates calcium flux. Cell. 1995;83:463–472. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
