rRNA-like sequences occur in diverse primary transcripts: implications for the control of gene expression
- PMID: 9012798
- PMCID: PMC19527
- DOI: 10.1073/pnas.94.2.422
rRNA-like sequences occur in diverse primary transcripts: implications for the control of gene expression
Abstract
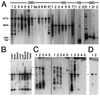
Many eukaryotic mRNAs contain sequences that resemble segments of 28S and 18S rRNAs, and these rRNA-like sequences are present in both the sense and antisense orientations. Some are similar to highly conserved regions of the rRNAs, whereas others have sequence similarities to expansion segments. In particular, four 18S rRNA-like sequences are found in several hundred different genes, and the location of these four sequences within the various genes is not random. One of these rRNA-like sequences is preferentially located within protein coding regions immediately upstream of the termination codon of a number of genes. Northern blot analysis of poly(A)+ RNA from different vertebrates (chicken, cattle, rat, mouse, and human) revealed that a large number of discrete RNA molecules hybridize at high stringency to cloned probes prepared from the 28S or 18S rRNA sequences that were found to match those in mRNAs. Inhibition of polymerase II activity, which prevents the synthesis of most mRNAs, abolished most of the hybridization to the rRNA probes. We consider the hypotheses that rRNA-like sequences may have spread throughout eukaryotic genomes and that their presence in primary transcripts may differentially affect gene expression.
Figures
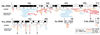
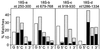

References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

