A point mutation leads to altered product specificity in beta-lactamase catalysis
- PMID: 9012802
- PMCID: PMC19531
- DOI: 10.1073/pnas.94.2.443
A point mutation leads to altered product specificity in beta-lactamase catalysis
Abstract
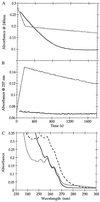
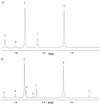
beta-Lactamases are the primary cause of beta-lactam antibiotic resistance in many pathogenic organisms. The beta-lactamase catalytic mechanism has been shown to involve a covalent acyl-enzyme. Examination of the structure of the class A beta-lactamase from Bacillus licheniformis suggested that replacement of Asn-170 by leucine would disrupt the deacylation reaction by displacing the hydrolytic water molecule. When N170L beta-lactamase was reacted with penicillins, a novel product was formed. We postulate that with leucine at position 170 the acyl-enzyme undergoes deacylation by an intramolecular rearrangement (rather than hydrolysis) to form a thiazolidine-oxazolinone as the initial product. The oxazolinone subsequently undergoes rapid breakdown leading to the formation of N-phenylacetylglycine and N-formylpenicillamine. This appears to be the first reported case where a point mutation leads to a change in enzyme mechanism resulting in a substantially altered product, effectively changing the product specificity of beta-lactamase into that of D-Ala-D-Ala-carboxypeptidase interacting with benzylpenicillin.
Figures
References
-
- Neu H C. Science. 1992;257:1064–1073. - PubMed
-
- Herzberg O, Moult J. Science. 1987;236:694–701. - PubMed
-
- Moews P C, Knox J R, Dideberg O, Charlier P, Frere J M. Protein Struct Funct Genet. 1990;7:156–171. - PubMed
-
- Strynadka N C J, Adachi H, Jensen S E, Johns K, Sielecki A, Betzel C, Sutoh K, James M N J. Nature (London) 1992;359:700–705. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

