The thioredoxin binding domain of bacteriophage T7 DNA polymerase confers processivity on Escherichia coli DNA polymerase I
- PMID: 9012809
- PMCID: PMC19538
- DOI: 10.1073/pnas.94.2.479
The thioredoxin binding domain of bacteriophage T7 DNA polymerase confers processivity on Escherichia coli DNA polymerase I
Abstract
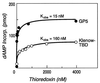
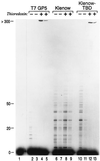
Bacteriophage T7 DNA polymerase shares extensive sequence homology with Escherichia coli DNA polymerase I. However, in vivo, E. coli DNA polymerase I is involved primarily in the repair of DNA whereas T7 DNA polymerase is responsible for the replication of the viral genome. In accord with these roles, T7 DNA polymerase is highly processive while E. coli DNA polymerase I has low processivity. The high processivity of T7 DNA polymerase is achieved through tight binding to its processivity factor, E. coli thioredoxin. We have identified a unique 76-residue domain in T7 DNA polymerase responsible for this interaction. Insertion of this domain into the homologous site in E. coli DNA polymerase I results in a dramatic increase in the processivity of the chimeric DNA polymerase, a phenomenon that is dependent upon its binding to thioredoxin.
Figures
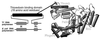

References
-
- Kornberg A, Baker T A. DNA Replication. 2nd Ed. New York: Freeman; 1992.
-
- Tabor S, Huber H E, Richardson C C. J Biol Chem. 1987;262:16212–16223. - PubMed
-
- Modrich P, Richardson C C. J Biol Chem. 1975;250:5515–5522. - PubMed
-
- Huber H E, Russel M, Model P, Richardson C C. J Biol Chem. 1986;261:15006–15012. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

