A novel membrane-associated metalloprotease, Ste24p, is required for the first step of NH2-terminal processing of the yeast a-factor precursor
- PMID: 9015299
- PMCID: PMC2134828
- DOI: 10.1083/jcb.136.2.271
A novel membrane-associated metalloprotease, Ste24p, is required for the first step of NH2-terminal processing of the yeast a-factor precursor
Abstract
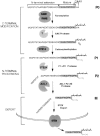
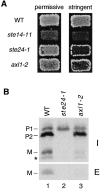
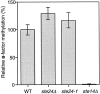
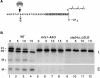
Many secreted bioactive signaling molecules, including the yeast mating pheromones a-factor and alpha-factor, are initially synthesized as precursors requiring multiple intracellular processing enzymes to generate their mature forms. To identify new gene products involved in the biogenesis of a-factor in Saccharomyces cerevisiae, we carried out a screen for MA Ta-specific, mating-defective mutants. We have identified a new mutant, ste24, in addition to previously known sterile mutants. During its biogenesis in a wild-type strain, the a-factor precursor undergoes a series of COOH-terminal CAAX modifications, two sequential NH2-terminal cleavage events, and export from the cell. Identification of the a-factor biosynthetic intermediate that accumulates in the ste24 mutant revealed that STE24 is required for the first NH2-terminal proteolytic processing event within the a-factor precursor, which takes place after COOH-terminal CAAX modification is complete. The STE24 gene product contains multiple predicted membrane spans, a zinc metalloprotease motif (HEXXH), and a COOH-terminal ER retrieval signal (KKXX). The HEXXH protease motif is critical for STE24 activity, since STE24 fails to function when conserved residues within this motif are mutated. The identification of Ste24p homologues in a diverse group of organisms, including Escherichia coli, Schizosaccharomyces pombe, Haemophilus influenzae, and Homo sapiens, indicates that Ste24p has been highly conserved throughout evolution. Ste24p and the proteins related to it define a new subfamily of proteins that are likely to function as intracellular, membrane-associated zinc metalloproteases.
Figures






References
-
- Adames N, Blundell K, Ashby MN, Boone C. Role of yeast insulin-degrading enzyme homologues in propheromone processing and bud site selection. Science (Wash DC) 1995;270:464–467. - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Anderegg RJ, Betz R, Carr SA, Crabb JW, Duntze W. Structure of Saccharomyces cerevisiae mating hormone a-factor. Identification of S-farnesyl cysteine as a structural component. J Biol Chem. 1988;263:18236–18240. - PubMed
-
- Ashby MN, Rine J. Ras and a-factor converting enzyme. Methods Enzymol. 1995;250:235–250. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

