Identification of a mid-anaphase checkpoint in budding yeast
- PMID: 9015305
- PMCID: PMC2134814
- DOI: 10.1083/jcb.136.2.345
Identification of a mid-anaphase checkpoint in budding yeast
Abstract
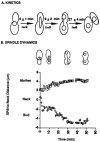
Activation of a facultative, dicentric chromosome provides a unique opportunity to introduce a double strand DNA break into a chromosome at mitosis. Time lapse video enhanced-differential interference contrast analysis of the cellular response upon dicentric activation reveals that the majority of cells initiates anaphase B, characterized by pole-pole separation, and pauses in mid-anaphase for 30-120 min with spindles spanning the neck of the bud before completing spindle elongation and cytokinesis. The length of the spindle at the delay point (3-4 microm) is not dependent on the physical distance between the two centromeres, indicating that the arrest represents surveillance of a dicentric induced aberration. No mid-anaphase delay is observed in the absence of the RAD9 checkpoint gene, which prevents cell cycle progression in the presence of damaged DNA. These observations reveal RAD9-dependent events well past the G2/M boundary and have considerable implications in understanding how chromosome integrity and the position and state of the mitotic spindle are monitored before cytokinesis.
Figures


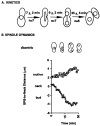
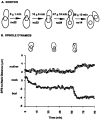
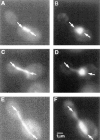


References
-
- Allen JB, Zhou Z, Siede W, Friedberg EC, Elledge SJ. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 1994;8:2401–2415. - PubMed
-
- Brock JA, Bloom K. A chromosome breakage assay to monitor mitotic forces in budding yeast. J Cell Sci. 1994;107:891–902. - PubMed
-
- Copeland CS, Snyder M. Nuclear pore complex antigens delineate nuclear envelope dynamics in vegetative and conjugating Saccharomyces cerevisiae. . Yeast. 1993;9:235–249. - PubMed
-
- Girard F, Fernandez A, Lamb N. Delayed cyclin A and B1 degradation in non-transformed mammalian cells. J Cell Sci. 1995;108:2599–2608. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

