Activation of the cell death program by inhibition of proteasome function
- PMID: 9023346
- PMCID: PMC19603
- DOI: 10.1073/pnas.94.3.855
Activation of the cell death program by inhibition of proteasome function
Abstract
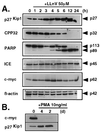
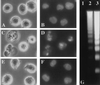
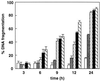
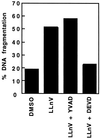
Activation of proteolytic enzymes, including cysteine proteases of the ced-3/ICE family, is a characteristic feature of the apoptotic program. In contrast, the role of the proteasome as the major nonlysosomal machinery to degrade or process proteins by ATP/ubiquitin-dependent proteolysis in this process is less clear. In human leukemic HL60 cells, inhibition of proteasome-mediated proteolysis by specific proteasomal inhibitors leads to the rapid induction of apoptosis as judged by morphological changes as well as by nuclear condensation and DNA fragmentation. HL60 apoptosis is due to activation of CPP32, a member of the ced-3/ICE family of cysteine proteases, and appears to occur independently from ICE activity. HL60 apoptosis is accompanied by an increase in the concentration of the cyclin-dependent kinase inhibitor p27Kip1. Labeling of the cells by the TUNEL technique demonstrates that HL60 cells undergoing apoptosis are primarily in the G1 phase of the cell cycle. Proteasomal activity therefore appears to be required in proliferating, but not in quiescent, HL60 cells for cell survival as well as normal progression through the cell cycle.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

