DNA double-strand-break sensitivity, DNA replication, and cell cycle arrest phenotypes of Ku-deficient Saccharomyces cerevisiae
- PMID: 9023348
- PMCID: PMC19605
- DOI: 10.1073/pnas.94.3.867
DNA double-strand-break sensitivity, DNA replication, and cell cycle arrest phenotypes of Ku-deficient Saccharomyces cerevisiae
Abstract
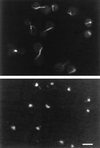
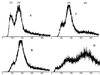
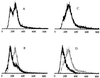
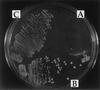
In mammalian cells, the Ku heterodimer is involved in DNA double-strand-break recognition and repair. We have established in yeast a connection between Ku activity and DNA double-strand-break damage repair, and a connection between Ku activity and commitment to DNA replication. We generated double-stranded DNA breaks in yeast cells in vivo by expressing a restriction endonuclease and have shown that yeast mutants lacking Ku p70 activity died while isogenic wild-type cells did not. Moreover, we have discovered that DNA damage occurs spontaneously during normal yeast mitotic growth, and that Ku functions in repair of this damage. We also observed that mitotically growing Ku p70 mutants have an anomalously high DNA content, suggesting a role for Ku in regulation of DNA synthesis. Finally, we present evidence that Ku p70 function is conserved between yeast, Drosophila, and humans.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

