Site-directed mutations reveal long-range compensatory interactions in the Adh gene of Drosophila melanogaster
- PMID: 9023359
- PMCID: PMC19616
- DOI: 10.1073/pnas.94.3.928
Site-directed mutations reveal long-range compensatory interactions in the Adh gene of Drosophila melanogaster
Abstract
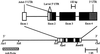
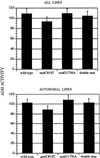
Long-range interactions between the 5' and 3' ends of mRNA molecules have been suggested to play a role in the initiation of translation and the regulation of gene expression. To identify such interactions and to study their molecular evolution, we used phylogenetic analysis to generate a model of mRNA higher-order structure in the Adh transcript of Drosophila melanogaster. This model predicts long-range, tertiary contacts between a region of the protein-encoding sequence just downstream of the start codon and a conserved sequence in the 3' untranslated region (UTR). To further examine the proposed structure, site-directed mutations were generated in vitro in a cloned D. melanogaster Adh gene, and the mutant constructs were introduced into the Drosophila germ line through P-element mediated transformation. Transformants were spectrophotometrically assayed for alcohol dehydrogenase activity. Our results indicate that transformants containing a silent mutation near the start of the protein-encoding sequence show an approximately 15% reduction in alcohol dehydrogenase activity relative to wild-type transformants. This activity can be restored to wild-type levels by a second, compensatory mutation in the 3' UTR. These observations are consistent with a higher-order structure model that includes long-range interactions between the 5' and 3' ends of the Adh mRNA. However, our results do not fit the classical compensatory substitution model because the second mutation by itself (in the 3' UTR) did not show a measurable reduction in gene expression.
Figures

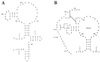
References
-
- Tarun S Z, Sachs A B. Genes Dev. 1995;9:2997–3007. - PubMed
-
- Dubnau J, Struhl G. Nature (London) 1996;379:694–699. - PubMed
-
- Rivera-Pomar R, Niessing D, Schmidt-Ott U, Gehring W J, Jäckle H. Nature (London) 1996;379:746–749. - PubMed
-
- Konings D A M, Van Duijn L P, Voorma H O, Hogeweg P. J Theor Biol. 1987;127:63–78. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

