Foreign (M13) DNA ingested by mice reaches peripheral leukocytes, spleen, and liver via the intestinal wall mucosa and can be covalently linked to mouse DNA
- PMID: 9023365
- PMCID: PMC19622
- DOI: 10.1073/pnas.94.3.961
Foreign (M13) DNA ingested by mice reaches peripheral leukocytes, spleen, and liver via the intestinal wall mucosa and can be covalently linked to mouse DNA
Abstract
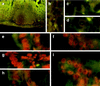
Food-ingested foreign DNA is not completely degraded in the gastrointestinal tract of mice. Phage M13mp18 DNA as a test molecule devoid of homology to mouse DNA was pipette-fed to or added to the food supply of mice. The fate of this foreign DNA in the animals was followed by several methods. In 84 animals, fragments of M13mp18 DNA were detected in the contents of the small intestine, the cecum (until 18 h), the large intestine, or the feces. In 254 animals, M13mp18 DNA fragments of up to 976 bp were found in blood 2-8 h after feeding. In buffer-fed control animals, M13mp18 DNA could not be detected. M13mp18 DNA fragments were traced by PCR in peripheral leukocytes and located by fluorescent in situ hybridization in about 1 of 1000 white cells between 2 and 8 h, and in spleen or liver cells up to 24 h after feeding, but not later. M13mp18 DNA could be traced by fluorescent in situ hybridization in the columnar epithelial cells, in the leukocytes in Peyer's patches of the cecum wall, in liver cells, and in B cells, T cells, and macrophages from spleen. These findings suggest transport of foreign DNA through the intestinal wall and Peyer's patches to peripheral blood leukocytes and into several organs. Upon extended feeding, M13mp18 DNA could be recloned from total spleen DNA into a lambda vector. Among about 2.5 x 10(7) lambda plaques, one plaque was isolated that contained a 1299 nucleotide pair fragment (nt 4736-6034) of sequence-identified M13mp18 DNA. This fragment was covalently linked to an 80 nt DNA segment with 70% homology to the mouse IgE receptor gene. The DNA from another lambda plaque also contained mouse DNA, bacterial DNA, and rearranged lambda DNA. Two additional plaques contained M13mp18 DNA fragments of at least 641 (nt 2660-3300) or 794 (nt 4640-5433) nucleotide pairs. The medical and evolutionary implications of these observations may be considerable.
Figures


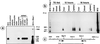

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

