Nested DNA inversion as a paradigm of programmed gene rearrangement
- PMID: 9023369
- PMCID: PMC19626
- DOI: 10.1073/pnas.94.3.985
Nested DNA inversion as a paradigm of programmed gene rearrangement
Abstract
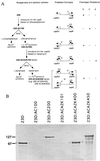
Programmed gene rearrangements are employed by a variety of microorganisms, including viruses, prokaryotes, and simple eukaryotes, to control gene expression. In most instances in which organisms mediate host evasion by large families of homologous gene cassettes, the mechanism of variation is not thought to involve DNA inversion. Here we report that Campylobacter fetus, a pathogenic Gram-negative bacterium, reassorts a single promoter, controlling surface-layer protein expression, and one or more complete ORFs strictly by DNA inversion. Rearrangements were independent of the distance between sites of inversion. These rearrangements permit variation in protein expression from the large surface-layer protein gene family and suggest an expanding paradigm of programmed DNA rearrangements among microorganisms.
Figures


References
-
- Borst P, Greaves D R. Science. 1982;235:658–667. - PubMed
-
- Beveridge T J, Koval S F. Advances in Paracrystalline Bacterial Surface Layers. New York: Plenum; 1993.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

