Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit
- PMID: 9023378
- PMCID: PMC19635
- DOI: 10.1073/pnas.94.3.1035
Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit
Abstract
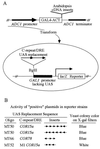
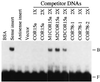
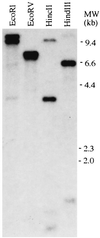
Recent efforts have defined a cis-acting DNA regulatory element in plants, the C-repeat/dehydration responsive element (DRE), that stimulates transcription in response to low temperature and water deficit. Here we report the isolation of an Arabidopsis thaliana cDNA that encodes a C-repeat/DRE binding factor, CBF1 (C-repeat/DRE Binding Factor 1). Analysis of the deduced CBF1 amino acid sequence indicates that the protein has a molecular mass of 24 kDa, a potential nuclear localization sequence, and a possible acidic activation domain. CBF1 also has an AP2 domain, which is a DNA-binding motif of about 60 aa present in the Arabidopsis proteins APETALA2, AINTEGUMENTA, and TINY; the tobacco ethylene response element binding proteins; and numerous other plant proteins of unknown function. The transcript levels for CBF1, which appears to be a single or low copy number gene, did not change appreciably in plants exposed to low temperature or in detached leaves subjected to water deficit. Binding of CBF1 to the C-repeat/DRE was demonstrated in gel shift assays using recombinant CBF1 protein expressed in Escherichia coli. Moreover, expression of CBF1 in yeast was found to activate transcription of reporter genes containing the C-repeat/DRE as an upstream activator sequence but not mutant versions of the DNA element. We conclude that CBF1 can function as a transcriptional activator that binds to the C-repeat/DRE DNA regulatory element and, thus, is likely to have a role in cold- and dehydration-regulated gene expression in Arabidopsis.
Figures



References
-
- Steponkus P L, Uemura M, Webb M S. In: Advances in Low Temperature Biology. Steponkus P L, editor. Vol. 2. London: JAI; 1993. pp. 211–312.
-
- Guy C, Haskell D, Neven L, Klein P, Smelser C. Planta. 1992;188:265–270. - PubMed
-
- Siminovitch D, Cloutier Y. Cryobiology. 1983;20:487–503. - PubMed
-
- Thomashow M F. In: Plant Responses to Cellular Dehydration During Environmental Stress. Close T L, Bray E A, editors. Rockville, MD: Am. Soc. Plant Physiol.; 1993. pp. 137–143.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

