Sleep and sleep regulation in normal and prion protein-deficient mice
- PMID: 9030645
- PMCID: PMC6573394
- DOI: 10.1523/JNEUROSCI.17-05-01869.1997
Sleep and sleep regulation in normal and prion protein-deficient mice
Abstract
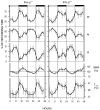
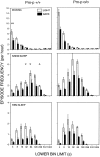
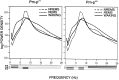
Mice are the preferred mammalian species for genetic investigations of the role of proteins. The normal function of the prion protein (PrP) is unknown, although it plays a major role in the prion diseases, including fatal familial insomnia. We investigated its role in sleep and sleep regulation by comparing baseline recordings and the effects of sleep deprivation in PrP knockout mice (129/SV) and wild-type controls (129/SV x C57BL/6), which are the mice used for most gene targeting experiments and whose behavior is not well characterized. Although no difference was evident in the amount of vigilance states, the null mice exhibited a larger degree of sleep fragmentation than the wild-type with almost double the amount of short waking episodes. As in other rodents, cortical temperature closely reflected the time course of waking. The increase of slow-wave activity (SWA; mean EEG power density in the 0.25-4.0 Hz range) at waking to nonrapid eye movement (NREM) sleep transitions was faster and reached a lower level in the null mice than in the wild-type. The contribution of the lower frequencies (0.25-5.0 Hz) to the spectrum was smaller than in other rodents in all three vigilance states, and the distinction between NREM sleep and REM sleep was most marked in the theta band. After the sleep deprivation, SWA was increased, but the changes in EEG power density and SWA were more prominent and lasted longer in the PrP-null mice. Our results suggest that PrP plays a role in promoting sleep continuity.
Figures




References
-
- Achermann P, Dijk DJ, Brunner DP, Borbély AA. A model of human sleep homeostasis based on EEG slow-wave activity: quantitative comparison of data and simulations. Brain Res Bull. 1993;31:97–113. - PubMed
-
- Aguzzi A, Weissmann C. Sleepless in Bologna: transmission of fatal familial insomnia. Trends Microbiol. 1996;4:129–131. - PubMed
-
- Aeschbach D, Cajochen C, Landolt HP, Borbély AA. Homeostatic sleep regulation in habitual short sleepers and long sleepers. Am J Physiol. 1996;270:41–53. - PubMed
-
- Borbély AA. Sleep homeostasis and models of sleep regulation. In: Kryger MH, Roth T, Dement WC, editors. Principles and practice of sleep medicine, 2nd Ed. Saunders; Philadelphia: 1994. pp. 309–320.
-
- Borbély AA, Baumann F, Brandeis D, Strauch I, Lehmann D. Sleep deprivation: effect on sleep stages and EEG power density in man. Electroencephalogr Clin Neurophysiol. 1981;51:483–493. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
