Characterization of mouse fibronectin alternative mRNAs reveals an unusual isoform present transiently during liver development
- PMID: 9041121
- PMCID: PMC6148308
Characterization of mouse fibronectin alternative mRNAs reveals an unusual isoform present transiently during liver development
Abstract
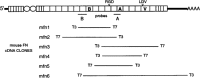
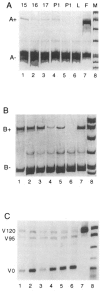
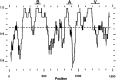
Fibronectins are found in many extracellular matrices as well as being abundant plasma proteins. The plasma isoforms of fibronectin, which are synthesized in the adult by liver hepatocytes, differ from those derived from most other cells and tissues due to alternative mRNA splicing. Studies in several vertebrates have indicated that FN alternative splicing is regulated spatially and temporally during development. The mouse represents an attractive organism in which to study the regulation of fibronectin splicing during development, but the patterns of fibronectin alternative splicing were not known for this species. Mouse fibronectin cDNA clones were isolated and sequenced, revealing > 95% identity with rat fibronectin at the amino acid level; all three segments that undergo alternative splicing are well conserved. RNase protection and RT-PCR were used to determine the patterns of alternative splicing that occur in fibroblasts and adult liver, sources of cellular and plasma fibronectins. Only A-B-mRNAs were detected in liver, and three V region variants were observed, corresponding to the protein isoforms V120, V95, and V0. Fibroblasts produced mRNAs that were heterogeneous for A and B splicing, but all RNAs contained V120. These patterns contrast with the embryonic form (B+A+V120). Characterization of fibronectin mRNAs from livers of fetal and newborn mice revealed that a significant level of B+ mRNA was present throughout late gestation, declining at birth. Little A+ mRNA was present, and the adult liver V region pattern was observed at all stages. Thus, fibronectin splicing changes during liver development are noncoordinate. One consequence of this temporal regulation is the transient synthesis of B+ mRNAs, including a novel isoform, B+A-V0.
Figures




Similar articles
-
Fibronectin mRNA alternative splicing is temporally and spatially regulated during chondrogenesis in vivo and in vitro.Dev Dyn. 1996 Jun;206(2):219-30. doi: 10.1002/(SICI)1097-0177(199606)206:2<219::AID-AJA11>3.0.CO;2-Y. Dev Dyn. 1996. PMID: 8725289
-
Characterization of the expression of the alternative splicing of the ED-A, ED-B and V regions of fibronectin mRNA in bovine ovarian follicles and corpora lutea.Reprod Fertil Dev. 1999;11(6):367-77. doi: 10.1071/rd99087. Reprod Fertil Dev. 1999. PMID: 10972305
-
Selective secretion of alternatively spliced fibronectin variants.J Cell Biol. 1989 Dec;109(6 Pt 2):3445-53. doi: 10.1083/jcb.109.6.3445. J Cell Biol. 1989. PMID: 2600138 Free PMC article.
-
Alternative splicing of fibronectin: three variants, three functions.Bioessays. 1991 Oct;13(10):527-33. doi: 10.1002/bies.950131006. Bioessays. 1991. PMID: 1755828 Review.
-
The fibronectin gene as a model for splicing and transcription studies.FASEB J. 1996 Feb;10(2):248-57. doi: 10.1096/fasebj.10.2.8641558. FASEB J. 1996. PMID: 8641558 Review.
Cited by
-
Hepatitis B virus-mediated changes of apolipoprotein mRNA abundance in cultured hepatoma cells.J Virol. 2003 May;77(9):5503-6. doi: 10.1128/jvi.77.9.5503-5506.2003. J Virol. 2003. PMID: 12692252 Free PMC article.
References
-
- Aota S.-I.; Nagai T.; Yamada K. M. Characterization of regions of fibronectin besides the arginine-glycine-aspartic acid sequence required for adhesive function of the cell-binding domain using site-directed mutagenesis. J. Biol. Chem. 266:15938–15943; 1991. - PubMed
-
- Barkalow F. J. B.; Schwarzbauer J. E. Localization of the major heparin-binding site in fibronectin. J. Biol. Chem. 266:7812–7818; 1991. - PubMed
-
- Bennett V. D.; Pallante K. M.; Adams S. L. The splicing pattern of fibronectin mRNA changes during chondrogenesis resulting in an unusual form of the mRNA in cartilage. J. Biol. Chem. 266:5918–5924; 1991. - PubMed
-
- Borsi L.; Balza E.; Allemanni G.; Zardi L. Differential expression of the fibronectin isoform containing the ED-B oncofetal domain in normal fibroblast cell lines originating from different tissues. Exp. Cell Res. 199:98–105; 1992. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
