Nucleosome-mediated synergism between transcription factors on the mouse mammary tumor virus promoter
- PMID: 9096316
- PMCID: PMC20292
- DOI: 10.1073/pnas.94.7.2885
Nucleosome-mediated synergism between transcription factors on the mouse mammary tumor virus promoter
Abstract
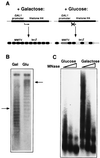
In unstimulated mammalian cells and in Saccharomyces cerevisiae, the mouse mammary tumor virus (MMTV) promoter is silent and organized into positioned nucleosomes, one of which encompasses the binding sites for glucocorticoid receptor (GR) and nuclear factor I (NFI). Glucocorticoid induction in vivo involves a functional synergism between GR and NFI and simultaneous occupancy of the promoter sites for both proteins that cannot be reproduced on naked DNA. The role of chromatin in the process of induction was investigated by manipulating the nucleosome density in yeast strains carrying a regulated histone H4 gene. Following depletion of nucleosomes, independent transactivation by NFI or by GR, as well as binding of the individual proteins to the MMTV promoter, were enhanced, in agreement with a repressive function of nucleosomes. In contrast, NFI-dependent hormone induction of the promoter and the simultaneous binding of receptor and NFI were compromised by nucleosome depletion. This effect could be partly mediated by a cryptic binding site for the receptor that is functional only in the nucleosomal context. Thus, positioned nucleosomes do not only account for constitutive repression of the MMTV promoter, but also participate in induction by mediating cooperative binding and functional synergism between GR and NFI.
Figures
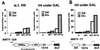



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

