Local DNA stretching mimics the distortion caused by the TATA box-binding protein
- PMID: 9096334
- PMCID: PMC20310
- DOI: 10.1073/pnas.94.7.2993
Local DNA stretching mimics the distortion caused by the TATA box-binding protein
Abstract
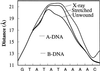
X-ray structures of the TATA box-binding protein complexed with its DNA target show that the nucleic acid is severely bent away from the protein and also strongly unwound. We have used molecular mechanics and energy mapping to understand how such an unusual conformation can be induced. The results show that simple deformation pathways involving local stretching or unwinding of DNA reproduce many features of the experimental structure. Notably, kinked junctions with the flanking B-DNA regions occur without the need for any specific local
Figures


References
-
- Kim Y, Gieger J H, Hahn S, Sigler P B. Nature (London) 1993;365:512–520. - PubMed
-
- Kim J L, Nikolov D B, Burley S K. Nature (London) 1993;365:520–527. - PubMed
-
- Kim J L, Burley S K. Nat Struct Biol. 1994;1:638–653. - PubMed
-
- Tan S, Hunziker Y, Sargent D F, Richmond T J. Nature (London) 1996;381:127–134. - PubMed
-
- Geiger J H, Hahn S, Lee S, Sigler P. Science. 1996;272:830–836. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

