A common mutational pattern in Cockayne syndrome patients from xeroderma pigmentosum group G: implications for a second XPG function
- PMID: 9096355
- PMCID: PMC20331
- DOI: 10.1073/pnas.94.7.3116
A common mutational pattern in Cockayne syndrome patients from xeroderma pigmentosum group G: implications for a second XPG function
Retraction in
-
Retraction for Nouspikel et al., A common mutational pattern in Cockayne syndrome patients from xeroderma pigmentosum group G: Implications for a second XPG function.Proc Natl Acad Sci U S A. 2006 Dec 19;103(51):19606. doi: 10.1073/pnas.0609759103. Epub 2006 Dec 13. Proc Natl Acad Sci U S A. 2006. PMID: 17179216 Free PMC article. No abstract available.
Abstract
Xeroderma pigmentosum (XP) patients have defects in nucleotide excision repair (NER), the versatile repair pathway that removes UV-induced damage and other bulky DNA adducts. Patients with Cockayne syndrome (CS), another rare sun-sensitive disorder, are specifically defective in the preferential removal of damage from the transcribed strand of active genes, a process known as transcription-coupled repair. These two disorders are usually clinically and genetically distinct, but complementation analyses have assigned a few CS patients to the rare XP groups B, D, or G. The XPG gene encodes a structure-specific endonuclease that nicks damaged DNA 3' to the lesion during NER. Here we show that three XPG/CS patients had mutations that would produce severely truncated XPG proteins. In contrast, two sibling XPG patients without CS are able to make full-length XPG, but with a missense mutation that inactivates its function in NER. These results suggest that XPG/CS mutations abolish interactions required for a second important XPG function and that it is the loss of this second function that leads to the CS clinical phenotype.
Figures
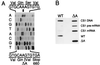

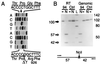



Comment in
-
Findings of scientific misconduct.NIH Guide Grants Contracts (Bethesda). 2006 Jun 9:NOT-OD-06-075. NIH Guide Grants Contracts (Bethesda). 2006. PMID: 16764107 Free PMC article. No abstract available.
-
Findings of Scientific Misconduct.Fed Regist. 2006 Jun 8;71(110):33308-33309. Fed Regist. 2006. PMID: 27737147 Free PMC article. No abstract available.
References
-
- Friedberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol.; 1995.
-
- Aboussekhra A, Biggerstaff M, Shivji M K K, Vilpo J A, Moncollin V, Podust V N, Protic M, Hübscher U, Egly J-M, Wood R D. Cell. 1995;80:859–868. - PubMed
-
- Mu D, Hsu D S, Sancar A. J Biol Chem. 1996;271:8285–8294. - PubMed
-
- Nance M A, Berry S A. Am J Med Genet. 1992;42:68–84. - PubMed
-
- Mellon I, Spivak G, Hanawalt P C. Cell. 1987;51:241–249. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

