Endosome to Golgi retrieval of the vacuolar protein sorting receptor, Vps10p, requires the function of the VPS29, VPS30, and VPS35 gene products
- PMID: 9105038
- PMCID: PMC2139870
- DOI: 10.1083/jcb.137.1.79
Endosome to Golgi retrieval of the vacuolar protein sorting receptor, Vps10p, requires the function of the VPS29, VPS30, and VPS35 gene products
Abstract
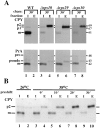
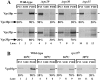
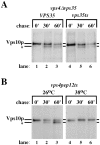
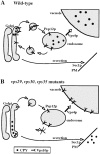
Mutations in the S. cerevisiae VPS29 and VPS30 genes lead to a selective protein sorting defect in which the vacuolar protein carboxypeptidase Y (CPY) is missorted and secreted from the cell, while other soluble vacuolar hydrolases like proteinase A (PrA) are delivered to the vacuole. This phenotype is similar to that seen in cells with mutations in the previously characterized VPS10 and VPS35 genes. Vps10p is a late Golgi transmembrane protein that acts as the sorting receptor for soluble vacuolar hydrolases like CPY and PrA, while Vps35p is a peripheral membrane protein which cofractionates with membranes enriched in Vps10p. The sequences of the VPS29, VPS30, and VPS35 genes do not yet give any clues to the functions of their products. Each is predicted to encode a hydrophilic protein with homologues in the human and C. elegans genomes. Interestingly, mutations in the VPS29, VPS30, or VPS35 genes change the subcellular distribution of the Vps10 protein, resulting in a shift of Vps10p from the Golgi to the vacuolar membrane. The route that Vps10p takes to reach the vacuole in a vps35 mutant does not depend upon Sec1p mediated arrival at the plasma membrane but does require the activity of the pre-vacuolar endosomal t-SNARE, Pep12p. A temperature conditional allele of the VPS35 gene was generated and has been found to cause missorting/secretion of CPY and also Vps10p to mislocalize to a vacuolar membrane fraction at the nonpermissive temperature. Vps35p continues to cofractionate with Vps10p in vps29 mutants, suggesting that Vps10p and Vps35p may directly interact. Together, the data indicate that the VPS29, VPS30, and VPS35 gene products are required for the normal recycling of Vps10p from the prevacuolar endosome back to the Golgi where it can initiate additional rounds of vacuolar hydrolase sorting.
Figures






References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

