Oat-maize chromosome addition lines: a new system for mapping the maize genome
- PMID: 9108009
- PMCID: PMC20472
- DOI: 10.1073/pnas.94.8.3524
Oat-maize chromosome addition lines: a new system for mapping the maize genome
Abstract
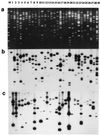
Novel plants with individual maize chromosomes added to a complete oat genome have been recovered via embryo rescue from oat (Avena sativa L., 2n = 6x = 42) x maize (Zea mays L., 2n = 20) crosses. An oat-maize disomic addition line possessing 21 pairs of oat chromosomes and one maize chromosome 9 pair was used to construct a cosmid library. A multiprobe (mixture of labeled fragments used as a probe) of highly repetitive maize-specific sequences was used to selectively isolate cosmid clones containing maize genomic DNA. Hybridization of individual maize cosmid clones or their subcloned fragments to maize and oat genomic DNA revealed that most high, middle, or low copy number DNA sequences are maize-specific. Such DNA markers allow the identification of maize genomic DNA in an oat genomic background. Chimeric cosmid clones were not found; apparently, significant exchanges of genetic material had not occurred between the maize-addition chromosome and the oat genome in these novel plants or in the cloning process. About 95% of clones selected at random from a maize genomic cosmid library could be detected by the multiprobe. The ability to selectively detect maize sequences in an oat background enables us to consider oat as a host for the cloning of specific maize chromosomes or maize chromosome segments. Introgressing maize chromosome segments into the oat genome via irradiation should allow the construction of a library of overlapping fragments for each maize chromosome to be used for developing a physical map of the maize genome.
Figures


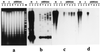
References
-
- Islam A K M R, Shepherd R, Sparrow D H B. Heredity. 1981;46:161–174.
-
- Riley R, Law C N. Stadler Genet Symp. 1984;16:301–322.
-
- Thomas H, Leggett J M, Jones I T. Euphytica. 1975;24:717–724.
-
- Jena K K, Khush G S. Genome. 1989;32:449–455.
-
- Jiang J, Morris K L, Gill B S. Chromosome Res. 1994;2:3–13. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

