Crystal structure of Lyme disease antigen outer surface protein A complexed with an Fab
- PMID: 9108020
- PMCID: PMC20483
- DOI: 10.1073/pnas.94.8.3584
Crystal structure of Lyme disease antigen outer surface protein A complexed with an Fab
Abstract
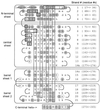
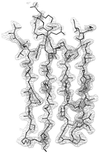
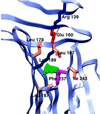
OspA (outer surface protein A) is an abundant immunogenic lipoprotein of the Lyme disease spirochete Borrelia burgdorferi. The crystal structure of a soluble recombinant form of OspA was solved in a complex with the Fab fragment of mouse monoclonal antibody 184.1 and refined to a resolution of 1.9 A. OspA has a repetitive antiparallel beta topology with an unusual nonglobular region of "freestanding" sheet connecting globular N- and C-terminal domains. Arrays of residues with alternating charges are a predominant feature of the folding pattern in the nonglobular region. The 184.1 epitope overlaps with a well conserved surface in the N-terminal domain, and a hydrophobic cavity buried in a positively charged cleft in the C-terminal domain is a potential binding site for an unknown ligand. An exposed variable region on the C-terminal domain of OspA is predicted to be an important factor in the worldwide effectiveness of OspA-based vaccines.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

