ELL2, a new member of an ELL family of RNA polymerase II elongation factors
- PMID: 9108030
- PMCID: PMC20493
- DOI: 10.1073/pnas.94.8.3639
ELL2, a new member of an ELL family of RNA polymerase II elongation factors
Abstract
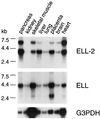
We recently isolated an RNA polymerase II elongation factor from rat liver nuclei and found it to be homologous to the product of the human ELL gene, a frequent target for translocations in acute myeloid leukemia. To further our understanding of the possible role(s) of ELL in transcriptional regulation and human disease, we initiated a search for ELL-related proteins. In this report we describe molecular cloning, expression, and characterization of human ELL2, a novel RNA polymerase II elongation factor 49% identical and 66% similar to ELL. Mechanistic studies indicate that ELL2 and ELL possess similar transcriptional activities. Structure-function studies localize the ELL2 elongation activation domain to an ELL2 N-terminal region that is highly homologous to ELL. Finally, Northern blot analysis reveals that the ELL2 and ELL genes are transcribed in many of the same tissues, but that the ratio of their transcripts exhibits tissue-to-tissue variation, raising the possibility that ELL2 and ELL may not perform completely general functions, but, instead, may perform gene- or tissue-specific functions.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

