Buried asparagines determine the dimerization specificities of leucine zipper mutants
- PMID: 9108036
- PMCID: PMC20499
- DOI: 10.1073/pnas.94.8.3673
Buried asparagines determine the dimerization specificities of leucine zipper mutants
Abstract
Regulation of gene expression by many transcription factors is controlled by specific combinations of homo- and heterodimers through a short alpha-helical coiled-coil known as a leucine zipper. The dimer interface of a leucine zipper involves side chains of the residues at the a, d, e, and g positions of the (abcdefg)n heptad repeat. To understand the basis for the specificity of dimer formation, we characterized GCN4 leucine zipper mutants with all 16 possible permutations and combinations of isoleucines and asparagines at four a positions in the dimer interface, using a genetic test for the specificity of dimer formation by lambda repressor-leucine zipper fusions. Heterodimers were detected by loss of repressor activity in the presence of a fusion to a dominant-negative mutant form of the DNA-binding domain of repressor. Reconstruction experiments using leucine zippers from GCN4, Jun, Fos, and C/EBP showed that this assay distinguishes pairs that form heterodimers from those that do not. We found that the mutants have novel dimerization specificities determined by the positioning of buried asparagine residues at the a positions. The pattern of buried polar residues could also explain the dimerization specificities of some naturally occurring leucine zippers. The altered specificity mutants described here should be useful for the construction of artificial regulatory circuitry.
Figures

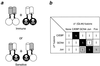

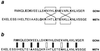
References
-
- O’Shea E K, Klemm J D, Kim P S, Alber T. Science. 1991;254:539–544. - PubMed
-
- Ellenberger T E, Brandl C J, Struhl K, Harrison S C. Cell. 1992;71:1223–1237. - PubMed
-
- Harbury P B, Zhang T, Kim P S, Alber T. Science. 1993;262:1401–1407. - PubMed
-
- Harbury P B, Kim P S, Alber T. Nature (London) 1994;371:80–83. - PubMed
-
- Glover J N M, Harrison S C. Nature (London) 1995;373:257–261. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

