Evolution of antifreeze glycoprotein gene from a trypsinogen gene in Antarctic notothenioid fish
- PMID: 9108060
- PMCID: PMC20523
- DOI: 10.1073/pnas.94.8.3811
Evolution of antifreeze glycoprotein gene from a trypsinogen gene in Antarctic notothenioid fish
Abstract
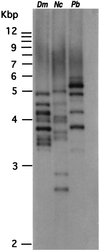
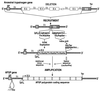
Freezing avoidance conferred by different types of antifreeze proteins in various polar and subpolar fishes represents a remarkable example of cold adaptation, but how these unique proteins arose is unknown. We have found that the antifreeze glycoproteins (AFGPs) of the predominant Antarctic fish taxon, the notothenioids, evolved from a pancreatic trypsinogen. We have determined the likely evolutionary process by which this occurred through characterization and analyses of notothenioid AFGP and trypsinogen genes. The primordial AFGP gene apparently arose through recruitment of the 5' and 3' ends of an ancestral trypsinogen gene, which provided the secretory signal and the 3' untranslated region, respectively, plus de novo amplification of a 9-nt Thr-Ala-Ala coding element from the trypsinogen progenitor to create a new protein coding region for the repetitive tripeptide backbone of the antifreeze protein. The small sequence divergence (4-7%) between notothenioid AFGP and trypsinogen genes indicates that the transformation of the proteinase gene into the novel ice-binding protein gene occurred quite recently, about 5-14 million years ago (mya), which is highly consistent with the estimated times of the freezing of the Antarctic Ocean at 10-14 mya, and of the main phyletic divergence of the AFGP-bearing notothenioid families at 7-15 mya. The notothenioid trypsinogen to AFGP conversion is the first clear example of how an old protein gene spawned a new gene for an entirely new protein with a new function. It also represents a rare instance in which protein evolution, organismal adaptation, and environmental conditions can be linked directly.
Figures


References
-
- Gon O, Heemstra P C. Fishes of the Southern Ocean. Grahamstown, South Africa: JLB Smith Institute of Ichthyology; 1990.
-
- Hubold G. In: Biology of Antarctic Fish. di Prisco G, Maresca M, Tota B, editors. Berlin: Springer; 1991. pp. 3–22.
-
- Eastman J T. Antarctic Fish Biology: Evolution in a Unique Environment. San Diego: Academic; 1993.
-
- DeWitt H H. In: Antarctic Map Folio Series, Folio 15. Bushnell V C, editor. New York: Am. Geogr. Soc.; 1971. pp. 1–10.
-
- Ekau W. Antarct Sci. 1990;2:129–137.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

